Autoantibody discovery across monogenic, acquired, and COVID-19-associated autoimmunity with scalable PhIP-seq
- PMID: 36300623
- PMCID: PMC9711525
- DOI: 10.7554/eLife.78550
Autoantibody discovery across monogenic, acquired, and COVID-19-associated autoimmunity with scalable PhIP-seq
Abstract
Phage immunoprecipitation sequencing (PhIP-seq) allows for unbiased, proteome-wide autoantibody discovery across a variety of disease settings, with identification of disease-specific autoantigens providing new insight into previously poorly understood forms of immune dysregulation. Despite several successful implementations of PhIP-seq for autoantigen discovery, including our previous work (Vazquez et al., 2020), current protocols are inherently difficult to scale to accommodate large cohorts of cases and importantly, healthy controls. Here, we develop and validate a high throughput extension of PhIP-seq in various etiologies of autoimmune and inflammatory diseases, including APS1, IPEX, RAG1/2 deficiency, Kawasaki disease (KD), multisystem inflammatory syndrome in children (MIS-C), and finally, mild and severe forms of COVID-19. We demonstrate that these scaled datasets enable machine-learning approaches that result in robust prediction of disease status, as well as the ability to detect both known and novel autoantigens, such as prodynorphin (PDYN) in APS1 patients, and intestinally expressed proteins BEST4 and BTNL8 in IPEX patients. Remarkably, BEST4 antibodies were also found in two patients with RAG1/2 deficiency, one of whom had very early onset IBD. Scaled PhIP-seq examination of both MIS-C and KD demonstrated rare, overlapping antigens, including CGNL1, as well as several strongly enriched putative pneumonia-associated antigens in severe COVID-19, including the endosomal protein EEA1. Together, scaled PhIP-seq provides a valuable tool for broadly assessing both rare and common autoantigen overlap between autoimmune diseases of varying origins and etiologies.
Keywords: APS1; COVID-19; IPEX; PhIP-seq; autoantibody; autoantigen; human; immunology; inflammation.
Conflict of interest statement
SV, SM, AB, AK, ZQ, EF, NL, DE, PB, SZ, JL, AM, IP, DY, CM, CW, BM, GS, KZ, AC, VT, CS, AT, KL, MW, OK, KD, OD, RB, LN, JB, JC, ML, TT, MA, JD No competing interests declared
Figures


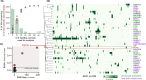
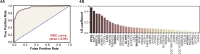
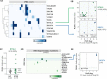



Update of
-
Autoantibody discovery across monogenic, acquired, and COVID19-associated autoimmunity with scalable PhIP-Seq.bioRxiv [Preprint]. 2022 Mar 24:2022.03.23.485509. doi: 10.1101/2022.03.23.485509. bioRxiv. 2022. Update in: Elife. 2022 Oct 27;11:e78550. doi: 10.7554/eLife.78550. PMID: 35350199 Free PMC article. Updated. Preprint.
Similar articles
-
Autoantibody discovery across monogenic, acquired, and COVID19-associated autoimmunity with scalable PhIP-Seq.bioRxiv [Preprint]. 2022 Mar 24:2022.03.23.485509. doi: 10.1101/2022.03.23.485509. bioRxiv. 2022. Update in: Elife. 2022 Oct 27;11:e78550. doi: 10.7554/eLife.78550. PMID: 35350199 Free PMC article. Updated. Preprint.
-
Exploring autoantigens in autoimmune limbic encephalitis using phage immunoprecipitation sequencing.J Neurol. 2025 Mar 25;272(4):292. doi: 10.1007/s00415-025-13039-7. J Neurol. 2025. PMID: 40133514
-
Validation of a murine proteome-wide phage display library for identification of autoantibody specificities.JCI Insight. 2023 Dec 8;8(23):e174976. doi: 10.1172/jci.insight.174976. JCI Insight. 2023. PMID: 37934865 Free PMC article.
-
Deciphering the autoreactome: Massively parallelized methods for autoantibody detection in humans.J Immunol Methods. 2025 Jun;541:113876. doi: 10.1016/j.jim.2025.113876. Epub 2025 May 6. J Immunol Methods. 2025. PMID: 40339788 Review.
-
Phage Immunoprecipitation and Sequencing-a Versatile Technique for Mapping the Antibody Reactome.Mol Cell Proteomics. 2024 Sep;23(9):100831. doi: 10.1016/j.mcpro.2024.100831. Epub 2024 Aug 19. Mol Cell Proteomics. 2024. PMID: 39168282 Free PMC article. Review.
Cited by
-
Phage Immunoprecipitation-Sequencing Reveals CDHR5 Autoantibodies in Select Patients With Interstitial Lung Disease.ACR Open Rheumatol. 2024 Sep;6(9):568-580. doi: 10.1002/acr2.11696. Epub 2024 Jul 1. ACR Open Rheumatol. 2024. PMID: 38952015 Free PMC article.
-
An autoantibody signature predictive for multiple sclerosis.Nat Med. 2024 May;30(5):1300-1308. doi: 10.1038/s41591-024-02938-3. Epub 2024 Apr 19. Nat Med. 2024. PMID: 38641750 Free PMC article.
-
The breadth of the neutralizing antibody response to original SARS-CoV-2 infection is linked to the presence of Long COVID symptoms.J Med Virol. 2023 Nov;95(11):e29216. doi: 10.1002/jmv.29216. J Med Virol. 2023. PMID: 37988251 Free PMC article.
-
Multiomics dissection of human RAG deficiency reveals distinctive patterns of immune dysregulation but a common inflammatory signature.Sci Immunol. 2025 Jan 10;10(103):eadq1697. doi: 10.1126/sciimmunol.adq1697. Epub 2025 Jan 10. Sci Immunol. 2025. PMID: 39792639
-
PhIP-Seq: methods, applications and challenges.Front Bioinform. 2024 Sep 4;4:1424202. doi: 10.3389/fbinf.2024.1424202. eCollection 2024. Front Bioinform. 2024. PMID: 39295784 Free PMC article. Review.
References
-
- Bacchetta R, Passerini L, Gambineri E, Dai M, Allan SE, Perroni L, Dagna-Bricarelli F, Sartirana C, Matthes-Martin S, Lawitschka A, Azzari C, Ziegler SF, Levings MK, Roncarolo MG. Defective regulatory and effector T cell functions in patients with FOXP3 mutations. The Journal of Clinical Investigation. 2006;116:1713–1722. doi: 10.1172/JCI25112. - DOI - PMC - PubMed
-
- Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann H-H, Zhang Y, Dorgham K, Philippot Q, Rosain J, Béziat V, Manry J, Shaw E, Haljasmägi L, Peterson P, Lorenzo L, Bizien L, Trouillet-Assant S, Dobbs K, de Jesus AA, Belot A, Kallaste A, Catherinot E, Tandjaoui-Lambiotte Y, Le Pen J, Kerner G, Bigio B, Seeleuthner Y, Yang R, Bolze A, Spaan AN, Delmonte OM, Abers MS, Aiuti A, Casari G, Lampasona V, Piemonti L, Ciceri F, Bilguvar K, Lifton RP, Vasse M, Smadja DM, Migaud M, Hadjadj J, Terrier B, Duffy D, Quintana-Murci L, van de Beek D, Roussel L, Vinh DC, Tangye SG, Haerynck F, Dalmau D, Martinez-Picado J, Brodin P, Nussenzweig MC, Boisson-Dupuis S, Rodríguez-Gallego C, Vogt G, Mogensen TH, Oler AJ, Gu J, Burbelo PD, Cohen JI, Biondi A, Bettini LR, D’Angio M, Bonfanti P, Rossignol P, Mayaux J, Rieux-Laucat F, Husebye ES, Fusco F, Ursini MV, Imberti L, Sottini A, Paghera S, Quiros-Roldan E, Rossi C, Castagnoli R, Montagna D, Licari A, Marseglia GL, Duval X, Ghosn J, HGID Lab. NIAID-USUHS Immune Response to COVID Group. COVID Clinicians. COVID-STORM Clinicians. Imagine COVID Group. French COVID Cohort Study Group. Milieu Intérieur Consortium. CoV-Contact Cohort. Amsterdam UMC Covid-19 Biobank. COVID Human Genetic Effort. Tsang JS, Goldbach-Mansky R, Kisand K, Lionakis MS, Puel A, Zhang S-Y, Holland SM, Gorochov G, Jouanguy E, Rice CM, Cobat A, Notarangelo LD, Abel L, Su HC, Casanova J-L. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585. doi: 10.1126/science.abd4585. - DOI - PMC - PubMed
-
- Bastard P, Gervais A, Le Voyer T, Rosain J, Philippot Q, Manry J, Michailidis E, Hoffmann HH, Eto S, Garcia-Prat M, Bizien L, Parra-Martínez A, Yang R, Haljasmägi L, Migaud M, Särekannu K, Maslovskaja J, de Prost N, Tandjaoui-Lambiotte Y, Luyt CE, Amador-Borrero B, Gaudet A, Poissy J, Morel P, Richard P, Cognasse F, Troya J, Trouillet-Assant S, Belot A, Saker K, Garçon P, Rivière JG, Lagier JC, Gentile S, Rosen LB, Shaw E, Morio T, Tanaka J, Dalmau D, Tharaux PL, Sene D, Stepanian A, Megarbane B, Triantafyllia V, Fekkar A, Heath JR, Franco JL, Anaya JM, Solé-Violán J, Imberti L, Biondi A, Bonfanti P, Castagnoli R, Delmonte OM, Zhang Y, Snow AL, Holland SM, Biggs C, Moncada-Vélez M, Arias AA, Lorenzo L, Boucherit S, Coulibaly B, Anglicheau D, Planas AM, Haerynck F, Duvlis S, Nussbaum RL, Ozcelik T, Keles S, Bousfiha AA, El Bakkouri J, Ramirez-Santana C, Paul S, Pan-Hammarström Q, Hammarström L, Dupont A, Kurolap A, Metz CN, Aiuti A, Casari G, Lampasona V, Ciceri F, Barreiros LA, Dominguez-Garrido E, Vidigal M, Zatz M, van de Beek D, Sahanic S, Tancevski I, Stepanovskyy Y, Boyarchuk O, Nukui Y, Tsumura M, Vidaur L, Tangye SG, Burrel S, Duffy D, Quintana-Murci L, Klocperk A, Kann NY, Shcherbina A, Lau YL, Leung D, Coulongeat M, Marlet J, Koning R, Reyes LF, Chauvineau-Grenier A, Venet F, Monneret G, Nussenzweig MC, Arrestier R, Boudhabhay I, Baris-Feldman H, Hagin D, Wauters J, Meyts I, Dyer AH, Kennelly SP, Bourke NM, Halwani R, Sharif-Askari NS, Dorgham K, Sallette J, Sedkaoui SM, AlKhater S, Rigo-Bonnin R, Morandeira F, Roussel L, Vinh DC, Ostrowski SR, Condino-Neto A, Prando C, Bonradenko A, Spaan AN, Gilardin L, Fellay J, Lyonnet S, Bilguvar K, Lifton RP, Mane S, Anderson MS, Boisson B, Béziat V, Zhang SY, Vandreakos E, Hermine O, Pujol A, Peterson P, Mogensen TH, Rowen L, Mond J, Debette S, de Lamballerie X, Duval X, Mentré F, Zins M, Soler-Palacin P, Colobran R, Gorochov G, Solanich X, Susen S, Martinez-Picado J, Raoult D, Vasse M, Gregersen PK, Piemonti L, Rodríguez-Gallego C, Notarangelo LD, Su HC, Kisand K, Okada S, Puel A, Jouanguy E, Rice CM, Tiberghien P, Zhang Q, Cobat A, Abel L, Casanova JL, HGID Lab. COVID Clinicians. COVID-STORM Clinicians. NIAID Immune Response to COVID Group. NH-COVAIR Study Group. Danish CHGE. Danish Blood Donor Study. French COVID Cohort Study Group. Imagine COVID-Group. Milieu Intérieur Consortium. CoV-Contact Cohort. Amsterdam UMC Covid-19. Biobank Investigators. COVID Human Genetic Effort. CONSTANCES cohort. 3C-Dijon Study. Cerba Health-Care. Etablissement du Sang study group Autoantibodies neutralizing type I ifns are present in ~4% of uninfected individuals over 70 years old and account for ~20% of COVID-19 deaths. Science Immunology. 2021a;6:eabl4340. doi: 10.1126/sciimmunol.abl4340. - DOI - PMC - PubMed
-
- Bastard P, Michailidis E, Hoffmann HH, Chbihi M, Le Voyer T, Rosain J, Philippot Q, Seeleuthner Y, Gervais A, Materna M, de Oliveira PMN, Maia M, Dinis Ano Bom AP, Azamor T, Araújo da Conceição D, Goudouris E, Homma A, Slesak G, Schäfer J, Pulendran B, Miller JD, Huits R, Yang R, Rosen LB, Bizien L, Lorenzo L, Chrabieh M, Erazo LV, Rozenberg F, Jeljeli MM, Béziat V, Holland SM, Cobat A, Notarangelo LD, Su HC, Ahmed R, Puel A, Zhang SY, Abel L, Seligman SJ, Zhang Q, MacDonald MR, Jouanguy E, Rice CM, Casanova JL. Auto-antibodies to type I ifns can underlie adverse reactions to yellow fever live attenuated vaccine. The Journal of Experimental Medicine. 2021b;218:e20202486. doi: 10.1084/jem.20202486. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

