Ablation of endothelial Atg7 inhibits ischemia-induced angiogenesis by upregulating Stat1 that suppresses Hif1a expression
- PMID: 36300763
- PMCID: PMC10240988
- DOI: 10.1080/15548627.2022.2139920
Ablation of endothelial Atg7 inhibits ischemia-induced angiogenesis by upregulating Stat1 that suppresses Hif1a expression
Abstract
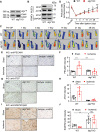
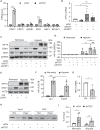
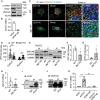
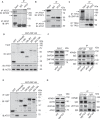
Ischemia-induced angiogenesis is critical for blood flow restoration and tissue regeneration, but the underlying molecular mechanism is not fully understood. ATG7 (autophagy related 7) is essential for classical degradative macroautophagy/autophagy and cell cycle regulation. However, whether and how ATG7 influences endothelial cell (EC) function and regulates post-ischemic angiogenesis remain unknown. Here, we showed that in mice subjected to femoral artery ligation, EC-specific deletion of Atg7 significantly impaired angiogenesis, delayed the recovery of blood flow reperfusion, and displayed reduction in HIF1A (hypoxia inducible factor 1 subunit alpha) expression. In addition, in cultured human umbilical vein endothelial cells (HUVECs), overexpression of HIF1A prevented ATG7 deficiency-reduced tube formation. Mechanistically, we identified STAT1 (signal transducer and activator of transcription 1) as a transcription suppressor of HIF1A and demonstrated that ablation of Atg7 upregulated STAT1 in an autophagy independent pathway, increased STAT1 binding to HIF1A promoter, and suppressed HIF1A expression. Moreover, lack of ATG7 in the cytoplasm disrupted the association between ATG7 and the transcription factor ZNF148/ZFP148/ZBP-89 (zinc finger protein 148) that is required for STAT1 constitutive expression, increased the binding between ZNF148/ZFP148/ZBP-89 and KPNB1 (karyopherin subunit beta 1), which promoted ZNF148/ZFP148/ZBP-89 nuclear translocation, and increased STAT1 expression. Finally, inhibition of STAT1 by fludarabine prevented the inhibition of HIF1A expression, angiogenesis, and blood flow recovery in atg7 KO mice. Our work reveals that lack of ATG7 inhibits angiogenesis by suppression of HIF1A expression through upregulation of STAT1 independently of autophagy under ischemic conditions, and suggest new therapeutic strategies for cancer and cardiovascular diseases.Abbreviations: ATG5: autophagy related 5; ATG7: autophagy related 7; atg7 KO: endothelial cell-specific atg7 knockout; BECN1: beclin 1; ChIP: chromatin immunoprecipitation; CQ: chloroquine; ECs: endothelial cells; EP300: E1A binding protein p300; HEK293: human embryonic kidney 293 cells; HIF1A: hypoxia inducible factor 1 subunit alpha; HUVECs: human umbilical vein endothelial cells; IFNG/IFN-γ: Interferon gamma; IRF9: interferon regulatory factor 9; KPNB1: karyopherin subunit beta 1; MAP1LC3A: microtubule associated protein 1 light chain 3 alpha; MEFs: mouse embryonic fibroblasts; MLECs: mouse lung endothelial cells; NAC: N-acetyl-l-cysteine; NFKB1/NFκB: nuclear factor kappa B subunit 1; PECAM1/CD31: platelet and endothelial cell adhesion molecule 1; RELA/p65: RELA proto-oncogene, NF-kB subunit; ROS: reactive oxygen species; SP1: Sp1 transcription factor; SQSTM1/p62: sequestosome 1; STAT1: signal transducer and activator of transcription 1; ULK1: unc-51 like autophagy activating kinase 1; ulk1 KO: endothelial cell-specific ulk1 knockout; VSMCs: mouse aortic smooth muscle cells; WT: wild type; ZNF148/ZFP148/ZBP-89: zinc finger protein 148.
Keywords: ATG7; Angiogenesis; HIF1A; STAT1; ZNF148/ZFP148/ZBP-89; endothelial cell; ischemia.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures





References
-
- Potente M, Gerhardt H, Carmeliet P.. Basic and therapeutic aspects of angiogenesis. Cell. 2011. Sep 16; 146(6):873–887. - PubMed
-
- Folkman J. Angiogenesis in cancer, vascular, rheumatoid and other disease. Nat Med. 1995. Jan;1(1):27–31. - PubMed
-
- Inampudi C, Akintoye E, Ando T, et al. Angiogenesis in peripheral arterial disease. Curr Opin Pharmacol. 2018. Apr;39:60–67. - PubMed
-
- Melincovici CS, Bosca AB, Susman S, et al. Vascular endothelial growth factor (VEGF) - key factor in normal and pathological angiogenesis. Rom J Morphol Embryol. 2018;59(2):455–467. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
