HIV skews the SARS-CoV-2 B cell response towards an extrafollicular maturation pathway
- PMID: 36300787
- PMCID: PMC9643005
- DOI: 10.7554/eLife.79924
HIV skews the SARS-CoV-2 B cell response towards an extrafollicular maturation pathway
Abstract
Background: HIV infection dysregulates the B cell compartment, affecting memory B cell formation and the antibody response to infection and vaccination. Understanding the B cell response to SARS-CoV-2 in people living with HIV (PLWH) may explain the increased morbidity, reduced vaccine efficacy, reduced clearance, and intra-host evolution of SARS-CoV-2 observed in some HIV-1 coinfections.
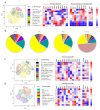
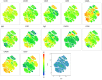
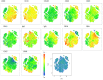
Methods: We compared B cell responses to COVID-19 in PLWH and HIV negative (HIV-ve) patients in a cohort recruited in Durban, South Africa, during the first pandemic wave in July 2020 using detailed flow cytometry phenotyping of longitudinal samples with markers of B cell maturation, homing, and regulatory features.
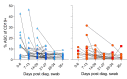
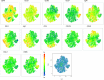
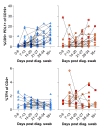
Results: This revealed a coordinated B cell response to COVID-19 that differed significantly between HIV-ve and PLWH. Memory B cells in PLWH displayed evidence of reduced germinal centre (GC) activity, homing capacity, and class-switching responses, with increased PD-L1 expression, and decreased Tfh frequency. This was mirrored by increased extrafollicular (EF) activity, with dynamic changes in activated double negative (DN2) and activated naïve B cells, which correlated with anti-RBD-titres in these individuals. An elevated SARS-CoV-2-specific EF response in PLWH was confirmed using viral spike and RBD bait proteins.
Conclusions: Despite similar disease severity, these trends were highest in participants with uncontrolled HIV, implicating HIV in driving these changes. EF B cell responses are rapid but give rise to lower affinity antibodies, less durable long-term memory, and reduced capacity to adapt to new variants. Further work is needed to determine the long-term effects of HIV on SARS-CoV-2 immunity, particularly as new variants emerge.
Funding: This work was supported by a grant from the Wellcome Trust to the Africa Health Research Institute (Wellcome Trust Strategic Core Award [grant number 201433/Z/16/Z]). Additional funding was received from the South African Department of Science and Innovation through the National Research Foundation (South African Research Chairs Initiative [grant number 64809]), and the Victor Daitz Foundation.
Keywords: B cell; COVID-19; HIV; RBD; SARS-CoV-2; epidemiology; global health; human; immunology; inflammation; spike.
© 2022, Krause et al.
Conflict of interest statement
RK, JS, HS, DM, FK, YG, AN, YZ, IG, MB, KK, MM, NM, DR, OL, JJ, DS, IW, AS, HK, TN, AL No competing interests declared, WH Reviewing editor, eLife
Figures








References
-
- Austin JW, Buckner CM, Kardava L, Wang W, Zhang X, Melson VA, Swanson RG, Martins AJ, Zhou JQ, Hoehn KB, Fisk JN, Dimopoulos Y, Chassiakos A, O’Dell S, Smelkinson MG, Seamon CA, Kwan RW, Sneller MC, Pittaluga S, Doria-Rose NA, McDermott A, Li Y, Chun T-W, Kleinstein SH, Tsang JS, Petrovas C, Moir S. Overexpression of T-bet in HIV infection is associated with accumulation of B cells outside germinal centers and poor affinity maturation. Science Translational Medicine. 2019;11:eaax0904. doi: 10.1126/scitranslmed.aax0904. - DOI - PMC - PubMed
-
- Bautista D, Vásquez C, Ayala-Ramírez P, Téllez-Sosa J, Godoy-Lozano E, Martínez-Barnetche J, Franco M, Angel J. Differential expression of igm and igd discriminates two subpopulations of human circulating igm+igd+CD27+ B cells that differ phenotypically, functionally, and genetically. Frontiers in Immunology. 2020;11:736. doi: 10.3389/fimmu.2020.00736. - DOI - PMC - PubMed
-
- Bhaskaran K, Rentsch CT, MacKenna B, Schultze A, Mehrkar A, Bates CJ, Eggo RM, Morton CE, Bacon SCJ, Inglesby P, Douglas IJ, Walker AJ, McDonald HI, Cockburn J, Williamson EJ, Evans D, Forbes HJ, Curtis HJ, Hulme WJ, Parry J, Hester F, Harper S, Evans SJW, Smeeth L, Goldacre B. HIV infection and COVID-19 death: a population-based cohort analysis of UK primary care data and linked national death registrations within the opensafely platform. The Lancet. HIV. 2021;8:e24–e32. doi: 10.1016/S2352-3018(20)30305-2. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

