Nucleoplasmic lamin C rapidly accumulates at sites of nuclear envelope rupture with BAF and cGAS
- PMID: 36301259
- PMCID: PMC9617480
- DOI: 10.1083/jcb.202201024
Nucleoplasmic lamin C rapidly accumulates at sites of nuclear envelope rupture with BAF and cGAS
Abstract
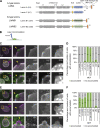
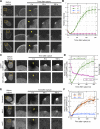
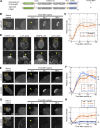
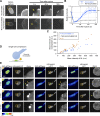
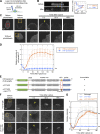
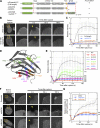
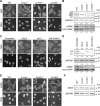
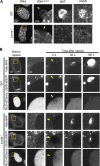
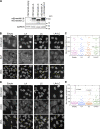
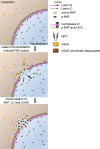
In mammalian cell nuclei, the nuclear lamina (NL) underlies the nuclear envelope (NE) to maintain nuclear structure. The nuclear lamins, the major structural components of the NL, are involved in the protection against NE rupture induced by mechanical stress. However, the specific role of the lamins in repair of NE ruptures has not been fully determined. Our analyses using immunofluorescence and live-cell imaging revealed that the nucleoplasmic pool of lamin C rapidly accumulated at sites of NE rupture induced by laser microirradiation in mouse embryonic fibroblasts. The accumulation of lamin C at the rupture sites required both the immunoglobulin-like fold domain that binds to barrier-to-autointegration factor (BAF) and a nuclear localization signal. The accumulation of nuclear BAF and cytoplasmic cyclic GMP-AMP synthase (cGAS) at the rupture sites was in part dependent on lamin A/C. These results suggest that nucleoplasmic lamin C, BAF, and cGAS concertedly accumulate at sites of NE rupture for rapid repair.
© 2022 Kono et al.
Figures







Similar articles
-
Mechanisms of A-Type Lamin Targeting to Nuclear Ruptures Are Disrupted in LMNA- and BANF1-Associated Progerias.Cells. 2022 Mar 2;11(5):865. doi: 10.3390/cells11050865. Cells. 2022. PMID: 35269487 Free PMC article.
-
Pervasive nuclear envelope ruptures precede ECM signaling and disease onset without activating cGAS-STING in Lamin-cardiomyopathy mice.Cell Rep. 2024 Jun 25;43(6):114284. doi: 10.1016/j.celrep.2024.114284. Epub 2024 May 29. Cell Rep. 2024. PMID: 38814785 Free PMC article.
-
The BAF A12T mutation disrupts lamin A/C interaction, impairing robust repair of nuclear envelope ruptures in Nestor-Guillermo progeria syndrome cells.Nucleic Acids Res. 2022 Sep 9;50(16):9260-9278. doi: 10.1093/nar/gkac726. Nucleic Acids Res. 2022. PMID: 36039758 Free PMC article.
-
Barrier-to-autointegration factor: a first responder for repair of nuclear ruptures.Cell Cycle. 2021 Apr;20(7):647-660. doi: 10.1080/15384101.2021.1892320. Epub 2021 Mar 8. Cell Cycle. 2021. PMID: 33678126 Free PMC article. Review.
-
Lamins in the nuclear interior - life outside the lamina.J Cell Sci. 2017 Jul 1;130(13):2087-2096. doi: 10.1242/jcs.203430. J Cell Sci. 2017. PMID: 28668931 Review.
Cited by
-
Crosstalk between mitotic reassembly and repair of the nuclear envelope.Nucleus. 2024 Dec;15(1):2352203. doi: 10.1080/19491034.2024.2352203. Epub 2024 May 23. Nucleus. 2024. PMID: 38780365 Free PMC article. Review.
-
cGAS: action in the nucleus.Front Immunol. 2024 Mar 7;15:1380517. doi: 10.3389/fimmu.2024.1380517. eCollection 2024. Front Immunol. 2024. PMID: 38515746 Free PMC article. Review.
-
VPS4B orchestrates response to nuclear envelope stress by regulating ESCRT-III dynamics in glioblastoma.Nucleus. 2024 Dec;15(1):2423660. doi: 10.1080/19491034.2024.2423660. Epub 2024 Nov 14. Nucleus. 2024. PMID: 39540606 Free PMC article.
-
Cryo-EM structures of the BAF-Lamin A/C complex bound to nucleosomes.Nat Commun. 2025 Feb 10;16(1):1495. doi: 10.1038/s41467-025-56823-9. Nat Commun. 2025. PMID: 39929866 Free PMC article.
-
A multiparametric anti-aging CRISPR screen uncovers a role for BAF in protein synthesis regulation.Nat Commun. 2025 Feb 16;16(1):1681. doi: 10.1038/s41467-025-56916-5. Nat Commun. 2025. PMID: 39956852 Free PMC article.
References
-
- Akinci, B., Onay H., Demir T., Savas-Erdeve S., Gen R., Simsir I.Y., Keskin F.E., Erturk M.S., Uzum A.K., Yaylali G.F., et al. 2017. Clinical presentations, metabolic abnormalities and end-organ complications in patients with familial partial lipodystrophy. Metabolism. 72:109–119. 10.1016/j.metabol.2017.04.010 - DOI - PubMed
-
- Béréziat, V., Cervera P., Le Dour C., Verpont M.C., Dumont S., Vantyghem M.C., Capeau J., Vigouroux C., and Lipodystrophy Study G.. 2011. LMNA mutations induce a non-inflammatory fibrosis and a brown fat-like dystrophy of enlarged cervical adipose tissue. Am. J. Pathol. 179:2443–2453. 10.1016/j.ajpath.2011.07.049 - DOI - PMC - PubMed
-
- Biamonti, G., Giacca M., Perini G., Contreas G., Zentilin L., Weighardt F., Guerra M., Della Valle G., Saccone S., Riva S., and Falaschi A.. 1992. The gene for a novel human lamin maps at a highly transcribed locus of chromosome 19 which replicates at the onset of S-phase. Mol. Cell. Biol. 12:3499–3506. 10.1128/mcb.12.8.3499-3506.1992 - DOI - PMC - PubMed
-
- Bonne, G., Di Barletta M.R., Varnous S., Bécane H.M., Hammouda E.H., Merlini L., Muntoni F., Greenberg C.R., Gary F., Urtizberea J.A., et al. 1999. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat. Genet. 21:285–288. 10.1038/6799 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

