An integrated SAGA and TFIID PIC assembly pathway selective for poised and induced promoters
- PMID: 36302553
- PMCID: PMC9732905
- DOI: 10.1101/gad.350026.122
An integrated SAGA and TFIID PIC assembly pathway selective for poised and induced promoters
Abstract
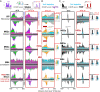
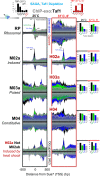
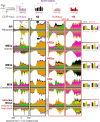
Genome-wide, little is understood about how proteins organize at inducible promoters before and after induction and to what extent inducible and constitutive architectures depend on cofactors. We report that sequence-specific transcription factors and their tethered cofactors (e.g., SAGA [Spt-Ada-Gcn5-acetyltransferase], Mediator, TUP, NuA4, SWI/SNF, and RPD3-L) are generally bound to promoters prior to induction ("poised"), rather than recruited upon induction, whereas induction recruits the preinitiation complex (PIC) to DNA. Through depletion and/or deletion experiments, we show that SAGA does not function at constitutive promoters, although a SAGA-independent Gcn5 acetylates +1 nucleosomes there. When inducible promoters are poised, SAGA catalyzes +1 nucleosome acetylation but not PIC assembly. When induced, SAGA catalyzes acetylation, deubiquitylation, and PIC assembly. Surprisingly, SAGA mediates induction by creating a PIC that allows TFIID (transcription factor II-D) to stably associate, rather than creating a completely TFIID-independent PIC, as generally thought. These findings suggest that inducible systems, where present, are integrated with constitutive systems.
Keywords: ChIP-seq; SAGA; Saccharomyces; gene regulation; genomics; transcription preinitiation complex.
© 2022 Mittal et al.; Published by Cold Spring Harbor Laboratory Press.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
