Miro GTPase domains regulate the assembly of the mitochondrial motor-adaptor complex
- PMID: 36302649
- PMCID: PMC9615026
- DOI: 10.26508/lsa.202201406
Miro GTPase domains regulate the assembly of the mitochondrial motor-adaptor complex
Abstract
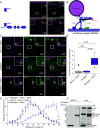
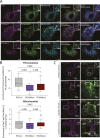
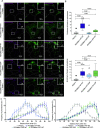
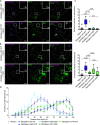
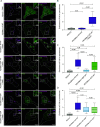
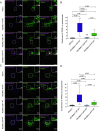
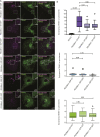
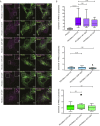
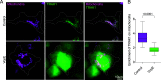
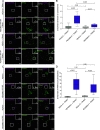
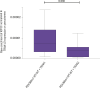
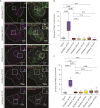
Mitochondrial transport relies on a motor-adaptor complex containing Miro1, a mitochondrial outer membrane protein with two GTPase domains, and TRAK1/2, kinesin-1, and dynein. Using a peroxisome-directed Miro1, we quantified the ability of GTPase mutations to influence the peroxisomal recruitment of complex components. Miro1 whose N-GTPase is locked in the GDP state does not recruit TRAK1/2, kinesin, or P135 to peroxisomes, whereas the GTP state does. Similarly, the expression of the MiroGAP VopE dislodges TRAK1 from mitochondria. Miro1 C-GTPase mutations have little influence on complex recruitment. Although Miro2 is thought to support mitochondrial motility, peroxisome-directed Miro2 did not recruit the other complex components regardless of the state of its GTPase domains. Neurons expressing peroxisomal Miro1 with the GTP-state form of the N-GTPase had markedly increased peroxisomal transport to growth cones, whereas the GDP-state caused their retention in the soma. Thus, the N-GTPase domain of Miro1 is critical for regulating Miro1's interaction with the other components of the motor-adaptor complex and thereby for regulating mitochondrial motility.
© 2022 Davis et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

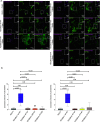

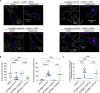


Similar articles
-
Interaction between the mitochondrial adaptor MIRO and the motor adaptor TRAK.J Biol Chem. 2023 Dec;299(12):105441. doi: 10.1016/j.jbc.2023.105441. Epub 2023 Nov 8. J Biol Chem. 2023. PMID: 37949220 Free PMC article.
-
Identification of Miro1 and Miro2 as mitochondrial receptors for myosin XIX.J Cell Sci. 2018 Sep 10;131(17):jcs219469. doi: 10.1242/jcs.219469. J Cell Sci. 2018. PMID: 30111583
-
Miro proteins coordinate microtubule- and actin-dependent mitochondrial transport and distribution.EMBO J. 2018 Feb 1;37(3):321-336. doi: 10.15252/embj.201696380. Epub 2018 Jan 8. EMBO J. 2018. PMID: 29311115 Free PMC article.
-
Mitochondrial trafficking in neurons and the role of the Miro family of GTPase proteins.Biochem Soc Trans. 2013 Dec;41(6):1525-31. doi: 10.1042/BST20130234. Biochem Soc Trans. 2013. PMID: 24256248 Review.
-
Miro sculpts mitochondrial dynamics in neuronal health and disease.Neurobiol Dis. 2016 Jun;90:27-34. doi: 10.1016/j.nbd.2015.12.008. Epub 2015 Dec 19. Neurobiol Dis. 2016. PMID: 26707701 Review.
Cited by
-
Structural-functional characterization of the MIRO1-TRAK1 complex.Nat Commun. 2025 Jul 4;16(1):6173. doi: 10.1038/s41467-025-61174-6. Nat Commun. 2025. PMID: 40615373 Free PMC article.
-
Interaction between the mitochondrial adaptor MIRO and the motor adaptor TRAK.J Biol Chem. 2023 Dec;299(12):105441. doi: 10.1016/j.jbc.2023.105441. Epub 2023 Nov 8. J Biol Chem. 2023. PMID: 37949220 Free PMC article.
-
Highly Specialized Mechanisms for Mitochondrial Transport in Neurons: From Intracellular Mobility to Intercellular Transfer of Mitochondria.Biomolecules. 2023 Jun 3;13(6):938. doi: 10.3390/biom13060938. Biomolecules. 2023. PMID: 37371518 Free PMC article. Review.
-
MIRO-1 interacts with VDAC-1 to regulate mitochondrial membrane potential in Caenorhabditis elegans.EMBO Rep. 2023 Aug 3;24(8):e56297. doi: 10.15252/embr.202256297. Epub 2023 Jun 12. EMBO Rep. 2023. PMID: 37306041 Free PMC article.
-
The peroxisome: an update on mysteries 3.0.Histochem Cell Biol. 2024 Feb;161(2):99-132. doi: 10.1007/s00418-023-02259-5. Epub 2024 Jan 20. Histochem Cell Biol. 2024. PMID: 38244103 Free PMC article. Review.
References
-
- Bocanegra JL, Fujita BM, Melton NR, Cowan JM, Schinski EL, Tamir TY, Major MB, Quintero OA (2020) The MyMOMA domain of MYO19 encodes for distinct Miro-dependent and Miro-independent mechanisms of interaction with mitochondrial membranes. Cytoskeleton (Hoboken) 77: 149–166. 10.1002/cm.21560 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
