Direct cell-to-cell transfer in stressed tumor microenvironment aggravates tumorigenic or metastatic potential in pancreatic cancer
- PMID: 36302783
- PMCID: PMC9613679
- DOI: 10.1038/s41525-022-00333-w
Direct cell-to-cell transfer in stressed tumor microenvironment aggravates tumorigenic or metastatic potential in pancreatic cancer
Abstract
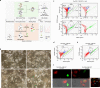
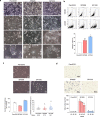
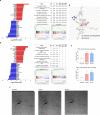
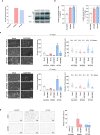
Pancreatic cancer exhibits a characteristic tumor microenvironment (TME) due to enhanced fibrosis and hypoxia and is particularly resistant to conventional chemotherapy. However, the molecular mechanisms underlying TME-associated treatment resistance in pancreatic cancer are not fully understood. Here, we developed an in vitro TME mimic system comprising pancreatic cancer cells, fibroblasts and immune cells, and a stress condition, including hypoxia and gemcitabine. Cells with high viability under stress showed evidence of increased direct cell-to-cell transfer of biomolecules. The resulting derivative cells (CD44high/SLC16A1high) were similar to cancer stem cell-like-cells (CSCs) with enhanced anchorage-independent growth or invasiveness and acquired metabolic reprogramming. Furthermore, CD24 was a determinant for transition between the tumorsphere formation or invasive properties. Pancreatic cancer patients with CD44low/SLC16A1low expression exhibited better prognoses compared to other groups. Our results suggest that crosstalk via direct cell-to-cell transfer of cellular components foster chemotherapy-induced tumor evolution and that targeting of CD44 and MCT1(encoded by SLC16A1) may be useful strategy to prevent recurrence of gemcitabine-exposed pancreatic cancers.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Microenvironmental networks promote tumor heterogeneity and enrich for metastatic cancer stem-like cells in Luminal-A breast tumor cells.Oncotarget. 2016 Dec 6;7(49):81123-81143. doi: 10.18632/oncotarget.13213. Oncotarget. 2016. PMID: 27835603 Free PMC article.
-
Epithelial mesenchymal transition correlates with CD24+CD44+ and CD133+ cells in pancreatic cancer.Oncol Rep. 2012 May;27(5):1599-605. doi: 10.3892/or.2012.1681. Epub 2012 Feb 7. Oncol Rep. 2012. PMID: 22322379
-
ALDH activity selectively defines an enhanced tumor-initiating cell population relative to CD133 expression in human pancreatic adenocarcinoma.PLoS One. 2011;6(6):e20636. doi: 10.1371/journal.pone.0020636. Epub 2011 Jun 13. PLoS One. 2011. PMID: 21695188 Free PMC article.
-
Cancer stem cells (CSCs) in cancer progression and therapy.J Cell Physiol. 2019 Jun;234(6):8381-8395. doi: 10.1002/jcp.27740. Epub 2018 Nov 11. J Cell Physiol. 2019. PMID: 30417375 Review.
-
Metabolic reprogramming due to hypoxia in pancreatic cancer: Implications for tumor formation, immunity, and more.Biomed Pharmacother. 2021 Sep;141:111798. doi: 10.1016/j.biopha.2021.111798. Epub 2021 Jun 10. Biomed Pharmacother. 2021. PMID: 34120068 Review.
Cited by
-
Transmembrane Protease Serine 11B Modulates Lactate Transport Through SLC16A1 in Pancreatic Ductal Adenocarcinoma-A Functional Link to Phenotype Heterogeneity.Int J Mol Sci. 2025 Jun 4;26(11):5398. doi: 10.3390/ijms26115398. Int J Mol Sci. 2025. PMID: 40508207 Free PMC article.
-
Deciphering cellular plasticity in pancreatic cancer for effective treatments.Cancer Metastasis Rev. 2024 Mar;43(1):393-408. doi: 10.1007/s10555-023-10164-5. Epub 2024 Jan 9. Cancer Metastasis Rev. 2024. PMID: 38194153 Review.
-
Microglia-mediated drug substance transfer promotes chemoresistance in brain tumors: insights from an in vitro co-culture model using GCV/Tk prodrug system.Cancer Cell Int. 2024 Jan 18;24(1):35. doi: 10.1186/s12935-024-03213-8. Cancer Cell Int. 2024. PMID: 38238749 Free PMC article.
-
Monocarboxylate Transporter-1 (MCT1)-Mediated Lactate Uptake Protects Pancreatic Adenocarcinoma Cells from Oxidative Stress during Glutamine Scarcity Thereby Promoting Resistance against Inhibitors of Glutamine Metabolism.Antioxidants (Basel). 2023 Sep 30;12(10):1818. doi: 10.3390/antiox12101818. Antioxidants (Basel). 2023. PMID: 37891897 Free PMC article.
-
The mosquito effect: regulatory and effector T cells acquire cytoplasmic material from tumor cells through intercellular transfer.Front Immunol. 2023 Dec 20;14:1272918. doi: 10.3389/fimmu.2023.1272918. eCollection 2023. Front Immunol. 2023. PMID: 38179041 Free PMC article.
References
-
- Gerlinger M. Targeted drugs ramp up cancer mutability. Science. 2019;366:1452–1453. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

