Targeting UHRF1-SAP30-MXD4 axis for leukemia initiating cell eradication in myeloid leukemia
- PMID: 36302855
- PMCID: PMC9715639
- DOI: 10.1038/s41422-022-00735-6
Targeting UHRF1-SAP30-MXD4 axis for leukemia initiating cell eradication in myeloid leukemia
Abstract
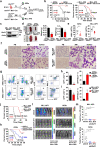
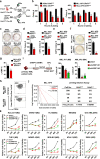
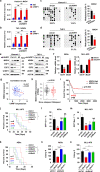
Aberrant self-renewal of leukemia initiation cells (LICs) drives aggressive acute myeloid leukemia (AML). Here, we report that UHRF1, an epigenetic regulator that recruits DNMT1 to methylate DNA, is highly expressed in AML and predicts poor prognosis. UHRF1 is required for myeloid leukemogenesis by maintaining self-renewal of LICs. Mechanistically, UHRF1 directly interacts with Sin3A-associated protein 30 (SAP30) through two critical amino acids, G572 and F573 in its SRA domain, to repress gene expression. Depletion of UHRF1 or SAP30 derepresses an important target gene, MXD4, which encodes a MYC antagonist, and leads to suppression of leukemogenesis. Further knockdown of MXD4 can rescue the leukemogenesis by activating the MYC pathway. Lastly, we identified a UHRF1 inhibitor, UF146, and demonstrated its significant therapeutic efficacy in the myeloid leukemia PDX model. Taken together, our study reveals the mechanisms for altered epigenetic programs in AML and provides a promising targeted therapeutic strategy against AML.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Howlader, N. et al. SEER Cancer Statistics Review(CSR) 1975–2016, 92 (National Cancer Institute, 2019).
-
- Jan, P. B., Maximilian, S. & Amer, M. Z. Are we witnessing the start of a therapeutic revolution in acute myeloid leukemia. Leuk. Lymphoma60, 1354–1369 (2019). - PubMed
-
- Rowley JD. Identificaton of a translocation with quinacrine fluorescence in a patient with acute leukemia. Ann. Genet. 1973;16:109–112. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

