SARS-CoV-2 Omicron variant is attenuated for replication in a polarized human lung epithelial cell model
- PMID: 36302956
- PMCID: PMC9610361
- DOI: 10.1038/s42003-022-04068-3
SARS-CoV-2 Omicron variant is attenuated for replication in a polarized human lung epithelial cell model
Abstract
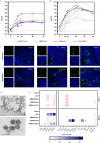
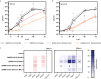
SARS-CoV-2 and its emerging variants of concern remain a major threat for global health. Here we introduce an infection model based upon polarized human Alveolar Epithelial Lentivirus immortalized (hAELVi) cells grown at the air-liquid interface to estimate replication and epidemic potential of respiratory viruses in the human lower respiratory tract. hAELVI cultures are highly permissive for different human coronaviruses and seasonal influenza A virus and upregulate various mediators following virus infection. Our analysis revealed a significantly reduced capacity of SARS-CoV-2 Omicron BA.1 and BA.2 variants to propagate in this human model compared to earlier D614G and Delta variants, which extends early risk assessments from epidemiological and animal studies suggesting a reduced pathogenicity of Omicron.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- WHO. Tracking SARS-CoV-2 Variants (WHO, 2021).
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

