Atypical nuclear envelope condensates linked to neurological disorders reveal nucleoporin-directed chaperone activities
- PMID: 36302970
- PMCID: PMC10041656
- DOI: 10.1038/s41556-022-01001-y
Atypical nuclear envelope condensates linked to neurological disorders reveal nucleoporin-directed chaperone activities
Abstract
DYT1 dystonia is a debilitating neurological movement disorder arising from mutation in the AAA+ ATPase TorsinA. The hallmark of Torsin dysfunction is nuclear envelope blebbing resulting from defects in nuclear pore complex biogenesis. Whether blebs actively contribute to disease manifestation is unknown. We report that FG-nucleoporins in the bleb lumen form aberrant condensates and contribute to DYT1 dystonia by provoking two proteotoxic insults. Short-lived ubiquitylated proteins that are normally rapidly degraded partition into the bleb lumen and become stabilized. In addition, blebs selectively sequester a specific HSP40-HSP70 chaperone network that is modulated by the bleb component MLF2. MLF2 suppresses the ectopic accumulation of FG-nucleoporins and modulates the selective properties and size of condensates in vitro. Our study identifies dual mechanisms of proteotoxicity in the context of condensate formation and establishes FG-nucleoporin-directed activities for a nuclear chaperone network.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests statement
The authors declare no competing interests.
Figures
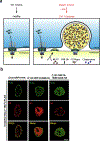







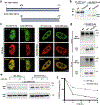



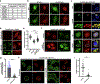

Comment in
-
Chaperoning the nuclear envelope.Nat Cell Biol. 2022 Nov;24(11):1563-1564. doi: 10.1038/s41556-022-01013-8. Nat Cell Biol. 2022. PMID: 36302972 No abstract available.
-
Nuclear Pore Complex Dysfunction in Dystonia Pathogenesis: Nucleoporins in the Spotlight.Mov Disord. 2023 Jan;38(1):23-24. doi: 10.1002/mds.29289. Epub 2022 Nov 24. Mov Disord. 2023. PMID: 36426380 No abstract available.
References
-
- Khan YA, White KI, and Brunger AT, The AAA+ superfamily: a review of the structural and mechanistic principles of these molecular machines. Crit Rev Biochem Mol Biol, 2021: p. 1–32. - PubMed
-
- Goodchild RE, Kim CE, and Dauer WT, Loss of the dystonia-associated protein torsinA selectively disrupts the neuronal nuclear envelope. Neuron, 2005. 48(6): p. 923–32. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- T32 GM007223/GM/NIGMS NIH HHS/United States
- R56MH122449/U.S. Department of Health & Human Services | NIH | National Institute of Mental Health (NIMH)
- R01GM114401/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- S10 OD023651/OD/NIH HHS/United States
- F31 NS120528/NS/NINDS NIH HHS/United States
- R01MH115939/U.S. Department of Health & Human Services | NIH | National Institute of Mental Health (NIMH)
- R01 MH115939/MH/NIMH NIH HHS/United States
- S10 OD019967/OD/NIH HHS/United States
- R01 NS105640/NS/NINDS NIH HHS/United States
- 5T32GM007223-44/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- F31MH116571/U.S. Department of Health & Human Services | NIH | National Institute of Mental Health (NIMH)
- S10 OD018034/OD/NIH HHS/United States
- F31NS120528/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- F31 MH116571/MH/NIMH NIH HHS/United States
- R56 MH122449/MH/NIMH NIH HHS/United States
- R01 GM114401/GM/NIGMS NIH HHS/United States
- NS105640/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

