Within-patient evolution of plasmid-mediated antimicrobial resistance
- PMID: 36303001
- PMCID: PMC7613874
- DOI: 10.1038/s41559-022-01908-7
Within-patient evolution of plasmid-mediated antimicrobial resistance
Abstract
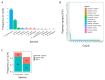
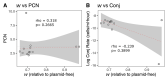
Antimicrobial resistance (AMR) in bacteria is a major threat to public health; one of the key elements in the spread and evolution of AMR in clinical pathogens is the transfer of conjugative plasmids. The drivers of AMR evolution have been studied extensively in vitro but the evolution of plasmid-mediated AMR in vivo remains poorly explored. Here, we tracked the evolution of the clinically relevant plasmid pOXA-48, which confers resistance to the last-resort antibiotics carbapenems, in a large collection of enterobacterial clones isolated from the gut of hospitalized patients. Combining genomic and experimental approaches, we first characterized plasmid diversity and the genotypic and phenotypic effects of multiple plasmid mutations on a common genetic background. Second, using cutting-edge genomic editing in wild-type multidrug-resistant enterobacteria, we dissected three cases of within-patient plasmid-mediated AMR evolution. Our results revealed compensatory evolution of plasmid-associated fitness cost and the evolution of enhanced plasmid-mediated AMR in bacteria evolving in the gut of hospitalized patients. Crucially, we observed that the evolution of pOXA-48-mediated AMR in vivo involves a pivotal trade-off between resistance levels and bacterial fitness. This study highlights the need to develop new evolution-informed approaches to tackle plasmid-mediated AMR dissemination.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures






Comment in
-
Plasmid evolution in the clinic.Nat Ecol Evol. 2022 Dec;6(12):1806-1807. doi: 10.1038/s41559-022-01907-8. Nat Ecol Evol. 2022. PMID: 36303002 No abstract available.
References
-
- Vincent J-L, et al. International study of the prevalence and outcomes of infection in intensive care units. JAMA. 2009;302:2323–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

