CRISPRi screens in human iPSC-derived astrocytes elucidate regulators of distinct inflammatory reactive states
- PMID: 36303069
- PMCID: PMC9633461
- DOI: 10.1038/s41593-022-01180-9
CRISPRi screens in human iPSC-derived astrocytes elucidate regulators of distinct inflammatory reactive states
Abstract
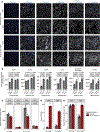
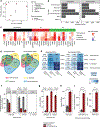
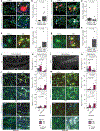
Astrocytes become reactive in response to insults to the central nervous system by adopting context-specific cellular signatures and outputs, but a systematic understanding of the underlying molecular mechanisms is lacking. In this study, we developed CRISPR interference screening in human induced pluripotent stem cell-derived astrocytes coupled to single-cell transcriptomics to systematically interrogate cytokine-induced inflammatory astrocyte reactivity. We found that autocrine-paracrine IL-6 and interferon signaling downstream of canonical NF-κB activation drove two distinct inflammatory reactive signatures, one promoted by STAT3 and the other inhibited by STAT3. These signatures overlapped with those observed in other experimental contexts, including mouse models, and their markers were upregulated in human brains in Alzheimer's disease and hypoxic-ischemic encephalopathy. Furthermore, we validated that markers of these signatures were regulated by STAT3 in vivo using a mouse model of neuroinflammation. These results and the platform that we established have the potential to guide the development of therapeutics to selectively modulate different aspects of inflammatory astrocyte reactivity.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
COMPETING INTERESTS STATEMENT
M. Kampmann is an inventor on US Patent 11,254,933 related to CRISPRi and CRISPRa screening, serves on the Scientific Advisory Boards of Engine Biosciences, Casma Therapeutics, Cajal Neuroscience, and Alector, and is an advisor to Modulo Bio and Recursion Therapeutics. J. TCW co-founded Asmos Therapeutics, LLC, serves on the scientific advisory board of NeuCyte, Inc, and has consulted for FIND Genomics Inc., CareCureSystems Corporation, TheWell Biosciences Inc., and Aleta Neuroscience, LLC. AG serves on the scientific advisory board for Genentech and is a consultant to Muna Therapeutics. None of the other authors declare competing interests.
Figures















References
-
- Hausmann ON Post-traumatic inflammation following spinal cord injury. Spinal Cord 41, 369–378 (2003). - PubMed
METHODS-ONLY REFERENCES
-
- TCW J, et al. Cholesterol and matrisome pathways dysregulated in human APOE ε4 glia. bioRxiv, 713362 (2019).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

