Adult human kidney organoids originate from CD24+ cells and represent an advanced model for adult polycystic kidney disease
- PMID: 36303074
- PMCID: PMC7613830
- DOI: 10.1038/s41588-022-01202-z
Adult human kidney organoids originate from CD24+ cells and represent an advanced model for adult polycystic kidney disease
Abstract
Adult kidney organoids have been described as strictly tubular epithelia and termed tubuloids. While the cellular origin of tubuloids has remained elusive, here we report that they originate from a distinct CD24+ epithelial subpopulation. Long-term-cultured CD24+ cell-derived tubuloids represent a functional human kidney tubule. We show that kidney tubuloids can be used to model the most common inherited kidney disease, namely autosomal dominant polycystic kidney disease (ADPKD), reconstituting the phenotypic hallmark of this disease with cyst formation. Single-cell RNA sequencing of CRISPR-Cas9 gene-edited PKD1- and PKD2-knockout tubuloids and human ADPKD and control tissue shows similarities in upregulation of disease-driving genes. Furthermore, in a proof of concept, we demonstrate that tolvaptan, the only approved drug for ADPKD, has a significant effect on cyst size in tubuloids but no effect on a pluripotent stem cell-derived model. Thus, tubuloids are derived from a tubular epithelial subpopulation and represent an advanced system for ADPKD disease modeling.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
All authors declare no competing interests. Disclosures: R.K. reports unrelated funding from Travere Therapeutics, Galapagos and Novo Nordisk and unrelated honoraria from Bayer, Pfizer, Grünenthal and Novo Nordisk. SH reports unrelated funding from Novo Nordisk. J.S.R. reports unrelated funding from GSK and Sanofi and unrelated fees from Travere Therapeutics and Astex Therapeutics. All other authors have nothing to disclose.
Figures

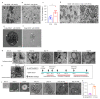






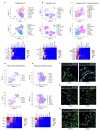


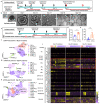
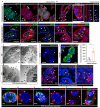


Comment in
-
Tubuloid as an alternative model of ADPKD.Kidney Int. 2023 Sep;104(3):409-411. doi: 10.1016/j.kint.2023.03.021. Epub 2023 Apr 5. Kidney Int. 2023. PMID: 37062357 No abstract available.
References
-
- Takasato M, et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature. 2015;526:564–568. - PubMed
-
- Taguchi A, et al. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell. 2014;14:53–67. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

