We are not ready yet: limitations of state-of-the-art disease named entity recognizers
- PMID: 36303237
- PMCID: PMC9612606
- DOI: 10.1186/s13326-022-00280-6
We are not ready yet: limitations of state-of-the-art disease named entity recognizers
Abstract
Background: Intense research has been done in the area of biomedical natural language processing. Since the breakthrough of transfer learning-based methods, BERT models are used in a variety of biomedical and clinical applications. For the available data sets, these models show excellent results - partly exceeding the inter-annotator agreements. However, biomedical named entity recognition applied on COVID-19 preprints shows a performance drop compared to the results on test data. The question arises how well trained models are able to predict on completely new data, i.e. to generalize.
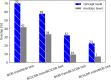
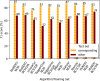
Results: Based on the example of disease named entity recognition, we investigate the robustness of different machine learning-based methods - thereof transfer learning - and show that current state-of-the-art methods work well for a given training and the corresponding test set but experience a significant lack of generalization when applying to new data.
Conclusions: We argue that there is a need for larger annotated data sets for training and testing. Therefore, we foresee the curation of further data sets and, moreover, the investigation of continual learning processes for machine learning-based models.
Keywords: BERT; Manual Curation; Text mining; bioNLP.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- School HM. N2C2: National NLP Clinical Challenges. https://n2c2.dbmi.hms.harvard.edu/. Accessed 20 June 2021.
-
- Doğan RI, Leaman R, Lu Z. The NCBI Disease Corpus. https://www.ncbi.nlm.nih.gov/CBBresearch/Dogan/DISEASE/. Accessed 11 July 2021.
-
- The NCBI Disease Corpus Guidelines. https://www.ncbi.nlm.nih.gov/CBBresearch/Dogan/DISEASE/Guidelines.html. Accessed 12 July 2021.
-
- The BC5CDR Corpus Guidelines. https://biocreative.bioinformatics.udel.edu/media/store/files/2015/bc5_C.... Accessed 12 July 2021.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

