The effect of repeat length on Marcal1-dependent single-strand annealing in Drosophila
- PMID: 36303322
- PMCID: PMC9836020
- DOI: 10.1093/genetics/iyac164
The effect of repeat length on Marcal1-dependent single-strand annealing in Drosophila
Abstract
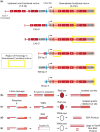
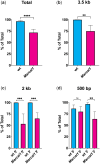
Proper repair of DNA double-strand breaks is essential to the maintenance of genomic stability and avoidance of genetic disease. Organisms have many ways of repairing double-strand breaks, including the use of homologous sequences through homology-directed repair. While homology-directed repair is often error free, in single-strand annealing homologous repeats flanking a double-strand break are annealed to one another, leading to the deletion of one repeat and the intervening sequences. Studies in yeast have shown a relationship between the length of the repeat and single-strand annealing efficacy. We sought to determine the effects of homology length on single-strand annealing in Drosophila, as Drosophila uses a different annealing enzyme (Marcal1) than yeast. Using an in vivo single-strand annealing assay, we show that 50 base pairs are insufficient to promote single-strand annealing and that 500-2,000 base pairs are required for maximum efficiency. Loss of Marcal1 generally followed the same homology length trend as wild-type flies, with single-strand annealing frequencies reduced to about a third of wild-type frequencies regardless of homology length. Interestingly, we find a difference in single-strand annealing rates between 500-base pair homologies that align to the annealing target either nearer or further from the double-strand break, a phenomenon that may be explained by Marcal1 dynamics. This study gives insights into Marcal1 function and provides important information to guide the design of genome engineering strategies that use single-strand annealing to integrate linear DNA constructs into a chromosomal double-strand break.
Keywords: DNA repair; DSB repair; annealing.
© The Author(s) 2022. Published by Oxford University Press on behalf of Genetics Society of America.
Conflict of interest statement
Conflicts of interest: None declared.
Figures


References
-
- Baradaran-Heravi A, Cho KS, Tolhuis B, Sanyal M, Morozova O, Morimoto M, Elizondo LI, Bridgewater D, Lubieniecka J, Beirnes K, et al. Penetrance of biallelic SMARCAL1 mutations is associated with environmental and genetic disturbances of gene expression. Hum Mol Genet. 2012;21(11):2572–2587. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

