Global population genetics and diversity in the TAS2R bitter taste receptor family
- PMID: 36303543
- PMCID: PMC9592824
- DOI: 10.3389/fgene.2022.952299
Global population genetics and diversity in the TAS2R bitter taste receptor family
Abstract
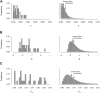
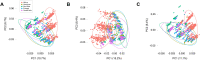
Bitter taste receptors (TAS2Rs) are noted for their role in perception, and mounting evidence suggests that they mediate responses to compounds entering airways, gut, and other tissues. The importance of these roles suggests that TAS2Rs have been under pressure from natural selection. To determine the extent of variation in TAS2Rs on a global scale and its implications for human evolution and behavior, we analyzed patterns of diversity in the complete 25 gene repertoire of human TAS2Rs in ∼2,500 subjects representing worldwide populations. Across the TAS2R family as a whole, we observed 721 single nucleotide polymorphisms (SNPs) including 494 nonsynonymous SNPs along with 40 indels and gained and lost start and stop codons. In addition, computational predictions identified 169 variants particularly likely to affect receptor function, making them candidate sources of phenotypic variation. Diversity levels ranged widely among loci, with the number of segregating sites ranging from 17 to 41 with a mean of 32 among genes and per nucleotide heterozygosity (π) ranging from 0.02% to 0.36% with a mean of 0.12%. F ST ranged from 0.01 to 0.26 with a mean of 0.13, pointing to modest differentiation among populations. Comparisons of observed π and F ST values with their genome wide distributions revealed that most fell between the 5th and 95th percentiles and were thus consistent with expectations. Further, tests for natural selection using Tajima's D statistic revealed only two loci departing from expectations given D's genome wide distribution. These patterns are consistent with an overall relaxation of selective pressure on TAS2Rs in the course of recent human evolution.
Keywords: evolution; genetics; perception; selection; taste.
Copyright © 2022 Wooding and Ramirez.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
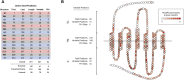
Similar articles
-
Worldwide haplotype diversity and coding sequence variation at human bitter taste receptor loci.Hum Mutat. 2005 Sep;26(3):199-204. doi: 10.1002/humu.20203. Hum Mutat. 2005. PMID: 16086309
-
Copy Number Variation in TAS2R Bitter Taste Receptor Genes: Structure, Origin, and Population Genetics.Chem Senses. 2016 Oct;41(8):649-59. doi: 10.1093/chemse/bjw067. Epub 2016 Jun 23. Chem Senses. 2016. PMID: 27340135 Free PMC article.
-
Tuning properties of avian and frog bitter taste receptors dynamically fit gene repertoire sizes.Mol Biol Evol. 2014 Dec;31(12):3216-27. doi: 10.1093/molbev/msu254. Epub 2014 Aug 31. Mol Biol Evol. 2014. PMID: 25180257
-
Bitter taste receptors: Genes, evolution and health.Evol Med Public Health. 2021 Oct 13;9(1):431-447. doi: 10.1093/emph/eoab031. eCollection 2021. Evol Med Public Health. 2021. PMID: 35154779 Free PMC article. Review.
-
Elucidation of mammalian bitter taste.Rev Physiol Biochem Pharmacol. 2005;154:37-72. doi: 10.1007/s10254-005-0041-0. Rev Physiol Biochem Pharmacol. 2005. PMID: 16032395 Review.
Cited by
-
Non-Volatile Compounds Involved in Bitterness and Astringency of Pulses: A Review.Molecules. 2023 Apr 7;28(8):3298. doi: 10.3390/molecules28083298. Molecules. 2023. PMID: 37110532 Free PMC article. Review.
-
Cross-ancestry analysis of brain QTLs enhances interpretation of schizophrenia genome-wide association studies.Am J Hum Genet. 2024 Nov 7;111(11):2444-2457. doi: 10.1016/j.ajhg.2024.09.001. Epub 2024 Oct 2. Am J Hum Genet. 2024. PMID: 39362218 Free PMC article.
-
Genomic Study of Taste Perception Genes in African Americans Reveals SNPs Linked to Alzheimer's Disease.bioRxiv [Preprint]. 2024 Aug 10:2024.08.10.607452. doi: 10.1101/2024.08.10.607452. bioRxiv. 2024. Update in: Sci Rep. 2024 Sep 16;14(1):21560. doi: 10.1038/s41598-024-71669-9. PMID: 39372803 Free PMC article. Updated. Preprint.
-
Bitter Taste Receptors in Bacterial Infections and Innate Immunity.Immun Inflamm Dis. 2025 Jul;13(7):e70232. doi: 10.1002/iid3.70232. Immun Inflamm Dis. 2025. PMID: 40709685 Free PMC article. Review.
-
The Remarkable Diversity of Vertebrate Bitter Taste Receptors: Recent Advances in Genomic and Functional Studies.Int J Mol Sci. 2024 Nov 25;25(23):12654. doi: 10.3390/ijms252312654. Int J Mol Sci. 2024. PMID: 39684366 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

