Potential of gut microbiota for lipopolysaccharide biosynthesis in European women with type 2 diabetes based on metagenome
- PMID: 36303603
- PMCID: PMC9592851
- DOI: 10.3389/fcell.2022.1027413
Potential of gut microbiota for lipopolysaccharide biosynthesis in European women with type 2 diabetes based on metagenome
Abstract
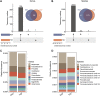
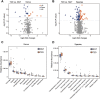
The abnormal accumulation of lipopolysaccharide (LPS) plays a crucial role in promoting type 2 diabetes (T2D). However, the capability of the gut microbiota to produce LPS in patients with T2D is still unclear, and evidence characterizing the patterns of gut microbiota with LPS productivity remains rare. This study aimed to uncover the profiles of LPS-biosynthesis-related enzymes and pathways, and explore the potential of LPS-producing gut microbiota in T2D. The gut metagenomic sequencing data from a European female cohort with normal glucose tolerance or untreated T2D were analyzed in this study. The sequence search revealed that the relative abundance of the critical enzymes responsible for LPS biosynthesis was significantly high in patients with T2D, especially for N-acetylglucosamine deacetylase, 3-deoxy-D-manno-octulosonic-acid transferase, and lauroyl-Kdo2-lipid IVA myristoyltransferase. The functional analysis indicated that a majority of pathways involved in LPS biosynthesis were augmented in patients with T2D. A total of 1,173 species from 335 genera containing the gene sequences of LPS enzymes, including LpxA/B/C/D/H/K/L/M and/or WaaA, coexisted in controls and patients with T2D. Critical taxonomies with discriminative fecal abundance between groups were revealed, which exhibited different associations with enzymes. Moreover, the identified gut microbial markers had correlations with LPS enzymes and were subsequently associated with microbial pathways. The present findings delineated the potential capability of gut microbiota toward LPS biosynthesis in European women and highlighted a gut microbiota-based mechanistic link between the disturbance in LPS biosynthesis and T2D. The restoration of LPS levels through gut microbiota manipulation might offer potential approaches for preventing and treating T2D.
Keywords: enzyme; gut microbiota; lipopolysaccharide; metagenome; type 2 diabetes.
Copyright © 2022 Dong, Wang, Yang, Chen and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Andreasen A. S., Pedersen-Skovsgaard T., Berg R. M., Svendsen K. D., Feldt-Rasmussen B., Pedersen B. K., et al. (2010). Type 2 diabetes mellitus is associated with impaired cytokine response and adhesion molecule expression in human endotoxemia. Intensive Care Med. 36, 1548–1555. 10.1007/s00134-010-1845-1 - DOI - PubMed
LinkOut - more resources
Full Text Sources

