Short Tandem Repeats as a High-Resolution Marker for Capturing Recent Orangutan Population Evolution
- PMID: 36303734
- PMCID: PMC9581056
- DOI: 10.3389/fbinf.2021.695784
Short Tandem Repeats as a High-Resolution Marker for Capturing Recent Orangutan Population Evolution
Abstract
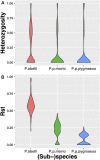
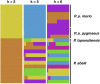
The genus Pongo is ideal to study population genetics adaptation, given its remarkable phenotypic divergence and the highly contrasting environmental conditions it's been exposed to. Studying its genetic variation bears the promise to reveal a motion picture of these great apes' evolutionary and adaptive history, and also helps us expand our knowledge of the patterns of adaptation and evolution. In this work, we advance the understanding of the genetic variation among wild orangutans through a genome-wide study of short tandem repeats (STRs). Their elevated mutation rate makes STRs ideal markers for the study of recent evolution within a given population. Current technological and algorithmic advances have rendered their sequencing and discovery more accurate, therefore their potential can be finally leveraged in population genetics studies. To study patterns of population variation within the wild orangutan population, we genotyped the short tandem repeats in a population of 21 individuals spanning four Sumatran and Bornean (sub-) species and eight Southeast Asian regions. We studied the impact of sequencing depth on our ability to genotype STRs and found that the STR copy number changes function as a powerful marker, correctly capturing the demographic history of these populations, even the divergences as recent as 10 Kya. Moreover, gene ontology enrichments for genes close to STR variants are aligned with local adaptations in the two islands. Coupled with more advanced STR-compatible population models, and selection tests, genomic studies based on STRs will be able to reduce the gap caused by the missing heritability for species with recent adaptations.
Keywords: genetic variation; local adaptation; orangutan; population diversity; recent evolution; short tandem repeats.
Copyright © 2021 Voicu, Krützen and Bilgin Sonay.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Miscellaneous

