Extending Association Rule Mining to Microbiome Pattern Analysis: Tools and Guidelines to Support Real Applications
- PMID: 36303759
- PMCID: PMC9580939
- DOI: 10.3389/fbinf.2021.794547
Extending Association Rule Mining to Microbiome Pattern Analysis: Tools and Guidelines to Support Real Applications
Abstract
Boosted by the exponential growth of microbiome-based studies, analyzing microbiome patterns is now a hot-topic, finding different fields of application. In particular, the use of machine learning techniques is increasing in microbiome studies, providing deep insights into microbial community composition. In this context, in order to investigate microbial patterns from 16S rRNA metabarcoding data, we explored the effectiveness of Association Rule Mining (ARM) technique, a supervised-machine learning procedure, to extract patterns (in this work, intended as groups of species or taxa) from microbiome data. ARM can generate huge amounts of data, making spurious information removal and visualizing results challenging. Our work sheds light on the strengths and weaknesses of pattern mining strategy into the study of microbial patterns, in particular from 16S rRNA microbiome datasets, applying ARM on real case studies and providing guidelines for future usage. Our results highlighted issues related to the type of input and the use of metadata in microbial pattern extraction, identifying the key steps that must be considered to apply ARM consciously on 16S rRNA microbiome data. To promote the use of ARM and the visualization of microbiome patterns, specifically, we developed microFIM (microbial Frequent Itemset Mining), a versatile Python tool that facilitates the use of ARM integrating common microbiome outputs, such as taxa tables. microFIM implements interest measures to remove spurious information and merges the results of ARM analysis with the common microbiome outputs, providing similar microbiome strategies that help scientists to integrate ARM in microbiome applications. With this work, we aimed at creating a bridge between microbial ecology researchers and ARM technique, making researchers aware about the strength and weaknesses of association rule mining approach.
Keywords: DNA metabarcoding; association rule mining; machine learning; microbiome data; microbiome patterns; pattern mining.
Copyright © 2022 Giulia, Anna, Antonia, Dario and Maurizio.
Conflict of interest statement
Author SA was employed by the company Quantia Consulting Srl. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
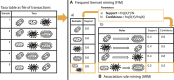
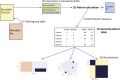

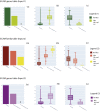

References
-
- Agrawal R., Imieliński T., Swami A. (1993). Mining Association Rules between Sets of Items in Large Databases. SIGMOD Rec. 22, 207–216. 10.1145/170036.170072 - DOI
-
- Agrawal R., Mannila H., Srikant R., Toivonen H., Verkamo A. I. (1996). Fast Discovery of Association Rules. Data Min. Knowl. Discov. 12 (1), 307–328.
-
- Anaconda Software Distribution (2020). Anaconda Documentation. Austin, TX, USA: Anaconda Inc. Available at: https://docs.anaconda.com/ .
LinkOut - more resources
Full Text Sources

