Predicted B Cell Epitopes Highlight the Potential for COVID-19 to Drive Self-Reactive Immunity
- PMID: 36303764
- PMCID: PMC9581003
- DOI: 10.3389/fbinf.2021.709533
Predicted B Cell Epitopes Highlight the Potential for COVID-19 to Drive Self-Reactive Immunity
Abstract
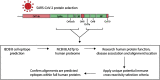
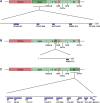
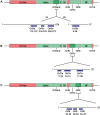
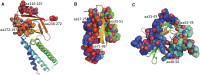
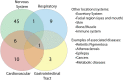
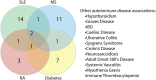
COVID-19, caused by the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), whilst commonly characterised as a respiratory disease, is reported to have extrapulmonary manifestations in multiple organs. Extrapulmonary involvement in COVID-19 includes autoimmune-like diseases such as Guillain-Barré syndrome and Kawasaki disease, as well as the presence of various autoantibodies including those associated with autoimmune diseases such a systemic lupus erythematosus (e.g. ANA, anti-La). Multiple strains of SARS-CoV-2 have emerged globally, some of which are found to be associated with increased transmissibility and severe disease. We performed an unbiased comprehensive mapping of the potential for cross-reactivity with self-antigens across multiple SARS-CoV-2 proteins and compared identified immunogenic regions across multiples strains. Using the Immune Epitope Database (IEDB) B cell epitope prediction tool, regions predicted as antibody epitopes with high prediction scores were selected. Epitope sequences were then blasted to eight other global strains to identify mutations within these regions. Of the 15 sequences compared, eight had a mutation in at least one other global strain. Predicted epitopes were then compared to human proteins using the NCBI blast tool. In contrast to studies focusing on short sequences of peptide identity, we have taken an immunological approach to selection criteria for further analysis and have identified 136 alignments of 6-23 amino acids (aa) in 129 human proteins that are immunologically likely to be cross-reactive with SARS-CoV-2. Additionally, to identify regions with significant potential to interfere with host cell function-or promote immunopathology, we identified epitope regions more likely to be accessible to pathogenic autoantibodies in the host, selected using a novel combination of sequence similarity, and modelling protein and alignment localization with a focus on extracellular regions. Our analysis identified 11 new predicted B-cell epitopes in host proteins, potentially capable of explaining key aspects of COVID-19 extrapulmonary pathology, and which were missed in other in silico studies which used direct identity rather than immunologically related functional criteria.
Keywords: COVID-19; autoimmunity; epitope mapping; molecular-mimicry; peptides; self-reactivity.
Copyright © 2021 Moody, Wilson, Boer, Holien, Flanagan, Jaworowski and Plebanski.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




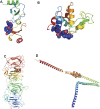
Similar articles
-
Using bioinformatic protein sequence similarity to investigate if SARS CoV-2 infection could cause an ocular autoimmune inflammatory reactions?Exp Eye Res. 2021 Feb;203:108433. doi: 10.1016/j.exer.2020.108433. Epub 2021 Jan 2. Exp Eye Res. 2021. PMID: 33400927 Free PMC article.
-
Immunoinformatics prediction of potential immunodominant epitopes from human coronaviruses and association with autoimmunity.Immunogenetics. 2022 Apr;74(2):213-229. doi: 10.1007/s00251-021-01250-5. Epub 2022 Jan 10. Immunogenetics. 2022. PMID: 35006282 Free PMC article.
-
Artificial intelligence predicts the immunogenic landscape of SARS-CoV-2 leading to universal blueprints for vaccine designs.Sci Rep. 2020 Dec 23;10(1):22375. doi: 10.1038/s41598-020-78758-5. Sci Rep. 2020. PMID: 33361777 Free PMC article.
-
Identification of B-Cell Epitopes for Eliciting Neutralizing Antibodies against the SARS-CoV-2 Spike Protein through Bioinformatics and Monoclonal Antibody Targeting.Int J Mol Sci. 2022 Apr 14;23(8):4341. doi: 10.3390/ijms23084341. Int J Mol Sci. 2022. PMID: 35457159 Free PMC article. Review.
-
Identification of Novel Candidate Epitopes on SARS-CoV-2 Proteins for South America: A Review of HLA Frequencies by Country.Front Immunol. 2020 Sep 3;11:2008. doi: 10.3389/fimmu.2020.02008. eCollection 2020. Front Immunol. 2020. PMID: 33013857 Free PMC article. Review.
Cited by
-
Autoantibodies in COVID-19 correlate with antiviral humoral responses and distinct immune signatures.Allergy. 2022 Aug;77(8):2415-2430. doi: 10.1111/all.15302. Epub 2022 Apr 8. Allergy. 2022. PMID: 35364615 Free PMC article.
-
The rise and spread of the SARS-CoV-2 AY.122 lineage in Russia.medRxiv [Preprint]. 2021 Dec 5:2021.12.02.21267168. doi: 10.1101/2021.12.02.21267168. medRxiv. 2021. Update in: Virus Evol. 2022 Mar 05;8(1):veac017. doi: 10.1093/ve/veac017. PMID: 34909799 Free PMC article. Updated. Preprint.
-
The rise and spread of the SARS-CoV-2 AY.122 lineage in Russia.Virus Evol. 2022 Mar 5;8(1):veac017. doi: 10.1093/ve/veac017. eCollection 2022. Virus Evol. 2022. PMID: 35371558 Free PMC article.
-
Annotation of the Extracellular Enveloped Form of Monkeypox Virus for the Design, Screening, Validation, and Simulation of a Chimeric Vaccine Construct.Biology (Basel). 2025 Jul 8;14(7):830. doi: 10.3390/biology14070830. Biology (Basel). 2025. PMID: 40723389 Free PMC article.
-
Contrasting Distribution of SARS-CoV-2 Lineages across Multiple Rounds of Pandemic Waves in West Bengal, the Gateway of East and North-East States of India.Microbiol Spectr. 2022 Aug 31;10(4):e0091422. doi: 10.1128/spectrum.00914-22. Epub 2022 Jul 19. Microbiol Spectr. 2022. PMID: 35852336 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

