Challenges in Bioinformatics Workflows for Processing Microbiome Omics Data at Scale
- PMID: 36303775
- PMCID: PMC9580927
- DOI: 10.3389/fbinf.2021.826370
Challenges in Bioinformatics Workflows for Processing Microbiome Omics Data at Scale
Abstract
The nascent field of microbiome science is transitioning from a descriptive approach of cataloging taxa and functions present in an environment to applying multi-omics methods to investigate microbiome dynamics and function. A large number of new tools and algorithms have been designed and used for very specific purposes on samples collected by individual investigators or groups. While these developments have been quite instructive, the ability to compare microbiome data generated by many groups of researchers is impeded by the lack of standardized application of bioinformatics methods. Additionally, there are few examples of broad bioinformatics workflows that can process metagenome, metatranscriptome, metaproteome and metabolomic data at scale, and no central hub that allows processing, or provides varied omics data that are findable, accessible, interoperable and reusable (FAIR). Here, we review some of the challenges that exist in analyzing omics data within the microbiome research sphere, and provide context on how the National Microbiome Data Collaborative has adopted a standardized and open access approach to address such challenges.
Keywords: bioinformatics; infrastructure; microbial ecology; microbiome; omics.
Copyright © 2022 Hu, Canon, Eloe-Fadrosh, Anubhav, Babinski, Corilo, Davenport, Duncan, Fagnan, Flynn, Foster, Hays, Huntemann, Jackson, Kelliher, Li, Lo, Mans, McCue, Mouncey, Mungall, Piehowski, Purvine, Smith, Varghese, Winston, Xu and Chain.
Conflict of interest statement
Author DW is employed by Polyneme LLC. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
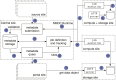


References
-
- Bundy J. G., Davey M. P., Viant M. R. (2008). Environmental Metabolomics: A Critical Review and Future Perspectives. Metabolomics 5 (1), 3–21. 10.1007/s11306-008-0152-0 - DOI

