New Approach to Accelerated Image Annotation by Leveraging Virtual Reality and Cloud Computing
- PMID: 36303792
- PMCID: PMC9580868
- DOI: 10.3389/fbinf.2021.777101
New Approach to Accelerated Image Annotation by Leveraging Virtual Reality and Cloud Computing
Abstract
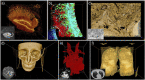
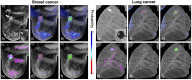
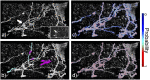
Three-dimensional imaging is at the core of medical imaging and is becoming a standard in biological research. As a result, there is an increasing need to visualize, analyze and interact with data in a natural three-dimensional context. By combining stereoscopy and motion tracking, commercial virtual reality (VR) headsets provide a solution to this critical visualization challenge by allowing users to view volumetric image stacks in a highly intuitive fashion. While optimizing the visualization and interaction process in VR remains an active topic, one of the most pressing issue is how to utilize VR for annotation and analysis of data. Annotating data is often a required step for training machine learning algorithms. For example, enhancing the ability to annotate complex three-dimensional data in biological research as newly acquired data may come in limited quantities. Similarly, medical data annotation is often time-consuming and requires expert knowledge to identify structures of interest correctly. Moreover, simultaneous data analysis and visualization in VR is computationally demanding. Here, we introduce a new procedure to visualize, interact, annotate and analyze data by combining VR with cloud computing. VR is leveraged to provide natural interactions with volumetric representations of experimental imaging data. In parallel, cloud computing performs costly computations to accelerate the data annotation with minimal input required from the user. We demonstrate multiple proof-of-concept applications of our approach on volumetric fluorescent microscopy images of mouse neurons and tumor or organ annotations in medical images.
Keywords: CT-scan; MRI; cloud computation; human-in-the-loop; inference; one-shot learning; virtual reality.
Copyright © 2022 Guérinot, Marcon, Godard, Blanc, Verdier, Planchon, Raimondi, Boddaert, Alonso, Sailor, Lledo, Hajj, El Beheiry and Masson.
Conflict of interest statement
MB and J-BM are cofounders, shareholders and, respectively, Chief Technology Officer (CTO) and Chief Scientific Officer (CSO) of AVATAR MEDICAL SAS, a startup that commercializes software for surgery planning in virtual reality. The DIVA software used in this study is not being commercialized by AVATAR MEDICAL SAS also the company’s technology is based on the same technology. The DIVA software which serves as base for this study is freely available and is reported in El Beheiry, et al. (2020). All developments within this paper are open source. HV was employed by Sanofi R&D. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Arivis A. G. (2021). Image Visualization and Analysis.
-
- Ayerbe V. M. C., Morales M. L. V., Rojas C. J. L., Cortés M. L. A. (2020). Visualization of 3d Models through Virtual Reality in the Planning of Congenital Cardiothoracic Anomalies Correction: An Initial Experience. World J. Pediatr. Congenit. Heart Surg. 11, 627–629. 10.1177/2150135120923618 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

