In silico discovery of biomarkers for the accurate and sensitive detection of Fusarium solani
- PMID: 36304265
- PMCID: PMC9580926
- DOI: 10.3389/fbinf.2022.972529
In silico discovery of biomarkers for the accurate and sensitive detection of Fusarium solani
Abstract
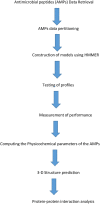
Fusarium solani is worrisome because it severely threatens the agricultural productivity of certain crops such as tomatoes and peas, causing the general decline, wilting, and root necrosis. It has also been implicated in the infection of the human eye cornea. It is believed that early detection of the fungus could save these crops from the destructive activities of the fungus through early biocontrol measures. Therefore, the present work aimed to build a sensitive model of novel anti-Fusarium solani antimicrobial peptides (AMPs) against the fungal cutinase 1 (CUT1) protein for early, sensitive and accurate detection. Fusarium solani CUT1 receptor protein 2D secondary structure, model validation, and functional motifs were predicted. Subsequently, anti-Fusarium solani AMPs were retrieved, and the HMMER in silico algorithm was used to construct a model of the AMPs. After their structure predictions, the interaction analysis was analyzed for the Fusarium solani CUT1 protein and the generated AMPs. The putative anti-Fusarium solani AMPs bound the CUT1 protein very tightly, with OOB4 having the highest binding energy potential for HDock. The pyDockWeb generated high electrostatic, desolvation, and low van der Waals energies for all the AMPs against CUT1 protein, with OOB1 having the most significant interaction. The results suggested the utilization of AMPs for the timely intervention, control, and management of these crops, as mentioned earlier, to improve their agricultural productivity and reduce their economic loss and the use of HMMER for constructing models for disease detection.
Keywords: Fusarium solani; energies; fungus; in silico; protein.
Copyright © 2022 Bakare, Gokul, Jimoh, Klein and Keyster.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Regulation of constitutively expressed and induced cutinase genes by different zinc finger transcription factors in Fusarium solani f. sp. pisi (Nectria haematococca).J Biol Chem. 2002 Mar 8;277(10):7905-12. doi: 10.1074/jbc.M108799200. Epub 2001 Dec 26. J Biol Chem. 2002. PMID: 11756444
-
PR-1-Like Protein as a Potential Target for the Identification of Fusarium oxysporum: An In Silico Approach.BioTech (Basel). 2021 Apr 14;10(2):8. doi: 10.3390/biotech10020008. BioTech (Basel). 2021. PMID: 35822818 Free PMC article.
-
Identification of biomarkers for the accurate and sensitive diagnosis of three bacterial pneumonia pathogens using in silico approaches.BMC Mol Cell Biol. 2020 Nov 20;21(1):82. doi: 10.1186/s12860-020-00328-4. BMC Mol Cell Biol. 2020. PMID: 33218302 Free PMC article.
-
Building HMM and molecular docking analysis for the sensitive detection of anti-viral pneumonia antimicrobial peptides (AMPs).Sci Rep. 2021 Oct 18;11(1):20621. doi: 10.1038/s41598-021-00223-8. Sci Rep. 2021. PMID: 34663864 Free PMC article.
-
The Fusarium solani species complex: ubiquitous pathogens of agricultural importance.Mol Plant Pathol. 2016 Feb;17(2):146-58. doi: 10.1111/mpp.12289. Epub 2015 Nov 4. Mol Plant Pathol. 2016. PMID: 26531837 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

