A Review of Feature Selection Methods for Machine Learning-Based Disease Risk Prediction
- PMID: 36304293
- PMCID: PMC9580915
- DOI: 10.3389/fbinf.2022.927312
A Review of Feature Selection Methods for Machine Learning-Based Disease Risk Prediction
Abstract
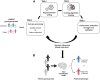
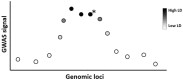
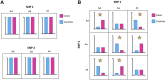
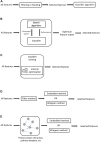
Machine learning has shown utility in detecting patterns within large, unstructured, and complex datasets. One of the promising applications of machine learning is in precision medicine, where disease risk is predicted using patient genetic data. However, creating an accurate prediction model based on genotype data remains challenging due to the so-called "curse of dimensionality" (i.e., extensively larger number of features compared to the number of samples). Therefore, the generalizability of machine learning models benefits from feature selection, which aims to extract only the most "informative" features and remove noisy "non-informative," irrelevant and redundant features. In this article, we provide a general overview of the different feature selection methods, their advantages, disadvantages, and use cases, focusing on the detection of relevant features (i.e., SNPs) for disease risk prediction.
Keywords: disease risk prediction; feature selection (FS); machine learing; risk prediction; statistical approaches.
Copyright © 2022 Pudjihartono, Fadason, Kempa-Liehr and O'Sullivan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Abramovich F., Benjamini Y., Donoho D. L., Johnstone I. M. (2006). Adapting to Unknown Sparsity by Controlling the False Discovery Rate. Ann. Stat. 34, 584–653. 10.1214/009053606000000074 - DOI
-
- Álvarez-Estévez D., Sánchez-Maroño N., Alonso-Betanzos A., Moret-Bonillo V. (2011). Reducing Dimensionality in a Database of Sleep EEG Arousals. Expert Syst. Appl. 38, 7746–7754.
-
- Alzubi R., Ramzan N., Alzoubi H., Amira A. (2017). A Hybrid Feature Selection Method for Complex Diseases SNPs. IEEE Access 6, 1292–1301. 10.1109/ACCESS.2017.2778268 - DOI
Publication types
LinkOut - more resources
Full Text Sources

