The Importance of Weakly Co-Evolving Residue Networks in Proteins is Revealed by Visual Analytics
- PMID: 36304294
- PMCID: PMC9580873
- DOI: 10.3389/fbinf.2022.836526
The Importance of Weakly Co-Evolving Residue Networks in Proteins is Revealed by Visual Analytics
Abstract
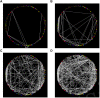
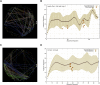
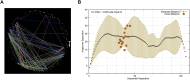
Small changes in a protein's core packing produce changes in function, and even small changes in function bias species fitness and survival. Therefore individually deleterious mutations should be evolutionarily coupled with compensating mutations that recover fitness. Co-evolving pairs of mutations should be littered across evolutionary history. Despite longstanding intuition, the results of co-evolution analyses have largely disappointed expectations. Regardless of the statistics applied, only a small majority of the most strongly co-evolving residues are typically found to be in contact, and much of the "meaning" of observed co-evolution has been opaque. In a medium-sized protein of 300 amino acids, there are almost 20 million potentially-important interdependencies. It is impossible to understand this data in textual format without extreme summarization or truncation. And, due to summarization and truncation, it is impossible to identify most patterns in the data. We developed a visualization approach that eschews the common "look at a long list of statistics" approach and instead enables the user to literally look at all of the co-evolution statistics simultaneously. Users of our tool reported visually obvious "clouds" of co-evolution statistics forming distinct patterns in the data, and analysis demonstrated that these clouds had structural relevance. To determine whether this phenomenon generalized, we repeated this experiment in three proteins we had not previously studied. The results provide evidence about how structural constrains have impacted co-evolution, why previous "examine the most frequently co-evolving residues" approaches have had limited success, and additionally shed light on the biophysical importance of different types of co-evolution.
Keywords: analytics; contact; correlations; evolution; proteins; structure; visualization.
Copyright © 2022 Mohan, Ozer and Ray.
Conflict of interest statement
HO was employed by the company Lilly Research Laboratories, Eli Lilly and Company. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Barral P., Batanero E., Palomares O., Quiralte J., Villalba M., Rodríguez R. (2004). A Major Allergen from Pollen Defines a Novel Family of Plant Proteins and Shows Intra- and Interspecies [correction of Interspecie] Cross-Reactivity. J. Immunol. 172, 3644–3651. 10.4049/jimmunol.172.6.3644 - DOI - PubMed
-
- Berry M. B., Phillips G. N., Jr. (1998). Crystal Structures of Bacillus Stearothermophilus Adenylate Kinase with Bound Ap5A, Mg2+ Ap5A, and Mn2+ Ap5A Reveal an Intermediate Lid Position and Six Coordinate Octahedral Geometry for Bound Mg2+ and Mn2+. Proteins 32, 276–288. 10.1002/(sici)1097-0134(19980815)32:3<276::aid-prot3>3.0.co;2-g - DOI - PubMed
LinkOut - more resources
Full Text Sources

