Rice Chalky Grain 5 regulates natural variation for grain quality under heat stress
- PMID: 36304400
- PMCID: PMC9593041
- DOI: 10.3389/fpls.2022.1026472
Rice Chalky Grain 5 regulates natural variation for grain quality under heat stress
Abstract
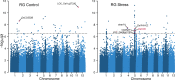
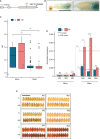
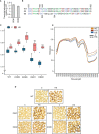
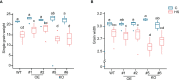
Heat stress occurring during rice (Oryza sativa) grain development reduces grain quality, which often manifests as increased grain chalkiness. Although the impact of heat stress on grain yield is well-studied, the genetic basis of rice grain quality under heat stress is less explored as quantifying grain quality is less tractable than grain yield. To address this, we used an image-based colorimetric assay (Red, R; and Green, G) for genome-wide association analysis to identify genetic loci underlying the phenotypic variation in rice grains exposed to heat stress. We found the R to G pixel ratio (RG) derived from mature grain images to be effective in distinguishing chalky grains from translucent grains derived from control (28/24°C) and heat stressed (36/32°C) plants. Our analysis yielded a novel gene, rice Chalky Grain 5 (OsCG5) that regulates natural variation for grain chalkiness under heat stress. OsCG5 encodes a grain-specific, expressed protein of unknown function. Accessions with lower transcript abundance of OsCG5 exhibit higher chalkiness, which correlates with higher RG values under stress. These findings are supported by increased chalkiness of OsCG5 knock-out (KO) mutants relative to wildtype (WT) under heat stress. Grains from plants overexpressing OsCG5 are less chalky than KOs but comparable to WT under heat stress. Compared to WT and OE, KO mutants exhibit greater heat sensitivity for grain size and weight relative to controls. Collectively, these results show that the natural variation at OsCG5 may contribute towards rice grain quality under heat stress.
Keywords: GWAS - genome-wide association study; chalkiness; grain; heat stress; imaging; rice.
Copyright © 2022 Chandran, Sandhu, Irvin, Paul, Dhatt, Hussain, Gao, Staswick, Yu, Morota and Walia.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Identifying Heat Adaptability QTLs and Candidate Genes for Grain Appearance Quality at the Flowering Stage in Rice.Rice (N Y). 2025 Mar 11;18(1):13. doi: 10.1186/s12284-025-00770-y. Rice (N Y). 2025. PMID: 40067644 Free PMC article.
-
Proteomic and Glycomic Characterization of Rice Chalky Grains Produced Under Moderate and High-temperature Conditions in Field System.Rice (N Y). 2016 Dec;9(1):26. doi: 10.1186/s12284-016-0100-y. Epub 2016 May 31. Rice (N Y). 2016. PMID: 27246013 Free PMC article.
-
High Temperature-Induced Expression of Rice α-Amylases in Developing Endosperm Produces Chalky Grains.Front Plant Sci. 2017 Dec 6;8:2089. doi: 10.3389/fpls.2017.02089. eCollection 2017. Front Plant Sci. 2017. PMID: 29270189 Free PMC article.
-
Relationship between High Temperature and Formation of Chalkiness and Their Effects on Quality of Rice.Biomed Res Int. 2018 Apr 1;2018:1653721. doi: 10.1155/2018/1653721. eCollection 2018. Biomed Res Int. 2018. PMID: 30065932 Free PMC article. Review.
-
The hot science in rice research: How rice plants cope with heat stress.Plant Cell Environ. 2023 Apr;46(4):1087-1103. doi: 10.1111/pce.14509. Epub 2022 Dec 19. Plant Cell Environ. 2023. PMID: 36478590 Review.
Cited by
-
Molecular Mechanisms of Grain Chalkiness Variation in Rice Panicles.Plants (Basel). 2025 Jan 16;14(2):244. doi: 10.3390/plants14020244. Plants (Basel). 2025. PMID: 39861596 Free PMC article.
-
Differential ABA sensitivity of superior and inferior rice grains is linked to cell cycle entry into endoreduplication.Front Plant Sci. 2025 May 20;16:1585022. doi: 10.3389/fpls.2025.1585022. eCollection 2025. Front Plant Sci. 2025. PMID: 40464007 Free PMC article.
-
Emerging strategies to improve heat stress tolerance in crops.aBIOTECH. 2025 Jan 24;6(1):97-115. doi: 10.1007/s42994-024-00195-z. eCollection 2025 Mar. aBIOTECH. 2025. PMID: 40060179 Free PMC article. Review.
-
Diversity of Gibberellin 2-oxidase genes in the barley genome offers opportunities for genetic improvement.J Adv Res. 2024 Dec;66:105-118. doi: 10.1016/j.jare.2023.12.021. Epub 2024 Jan 9. J Adv Res. 2024. PMID: 38199453 Free PMC article.
-
Identifying Heat Adaptability QTLs and Candidate Genes for Grain Appearance Quality at the Flowering Stage in Rice.Rice (N Y). 2025 Mar 11;18(1):13. doi: 10.1186/s12284-025-00770-y. Rice (N Y). 2025. PMID: 40067644 Free PMC article.
References
-
- Armstrong P. R., McClung A. M., Maghirang E. B., Chen M. H., Brabec D. L., Yaptenco K. F., et al. . (2019). Detection of chalk in single kernels of long-grain milled rice using imaging and visible/near-infrared instruments. Cereal Chem. 96, 1103–1111. doi: 10.1002/cche.10220 - DOI
-
- Baysal C., He W., Drapal M., Villorbina G., Medina V., Capell T., et al. . (2020). Inactivation of rice starch branching enzyme IIb triggers broad and unexpected changes in metabolism by transcriptional reprogramming. Proc. Natl. Acad. Sci. U. S. A. 117, 26503–26512. doi: 10.1073/pnas.2014860117 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

