CodY is modulated by YycF and affects biofilm formation in Staphylococcus aureus
- PMID: 36304951
- PMCID: PMC9593060
- DOI: 10.3389/fmicb.2022.967567
CodY is modulated by YycF and affects biofilm formation in Staphylococcus aureus
Abstract
Background: Staphylococcus aureus (S. aureus) is the leading cause of various infective diseases, including topical soft tissue infections. The goals of this study were to investigate the roles of YycF and CodY in the regulation of biofilm formation and pathogenicity.
Methods: Electrophoretic mobility shift assay (EMSA) was conducted to validate the bound promoter regions of YycF protein. We constructed the codY up-regulated or down-regulated S. aureus mutants. The biofilm biomass was determined by crystal violet microtiter assay and scanning electron microscopy (SEM). Quantitative RT-PCR analysis was used to detect the transcripts of biofilm-related genes. The live and dead cells of S. aureus biofilm were also investigated by confocal laser scanning microscopy (CLSM). We constructed an abscess infection in Sprague Dawley (SD) rat models to determine the effect of CodY on bacterial pathogenicity. We further used the RAW264.7, which were cocultured with S. aureus, to evaluate the effect of CodY on macrophages apoptosis.
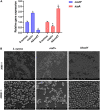
Result: Quantitative RT-PCR analyses reveled that YycF negatively regulates codY expression. EMSA assays indicated that YycF protein directly binds to the promoter regions of codY gene. Quantitative RT-PCR confirmed the construction of dual- mutant stains codY + ASyycF and codY-ASyycF. The SEM results showed that the biofilm formation in the codY + ASyycF group was sparser than those in the other groups. The crystal violet assays indicated that the codY + ASyycF group formed less biofilms, which was consistent with the immunofluorescence results of the lowest live cell ration in the codY + ASyycF group. The expression levels of biofilm-associated icaA gene were significantly reduced in the codY + strain, indicating codY negatively regulates the biofilm formation. Furthermore, CodY impedes the pathogenicity in a rat-infection model. After cocultured with bacteria or 4-h in vitro, the apoptosis rates of macrophage cells were lowest in the codY + group.
Conclusions: YycF negatively regulate the expression of codY. By interaction with codY, YycF could modulate S. aureus biofilm formation via both eDNA- dependent and PIA- dependent pathways, which can be a significant target for antibiofilm. CodY not only impedes the pathogenicity but also has a role on immunoregulation. Thus, the current evidence may provide a supplementary strategy for managing biofilm infections.
Keywords: CodY; Staphylococcus aureus; YycFG; antisense; biofilm formation.
Copyright © 2022 Wu, Qin, Deng, Liu, Zhang, Lei and Feng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
LinkOut - more resources
Full Text Sources

