Recruitment of distinct UDP-glycosyltransferase families demonstrates dynamic evolution of chemical defense within Eucalyptus L'Hér
- PMID: 36305250
- PMCID: PMC10107851
- DOI: 10.1111/nph.18581
Recruitment of distinct UDP-glycosyltransferase families demonstrates dynamic evolution of chemical defense within Eucalyptus L'Hér
Abstract
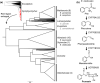
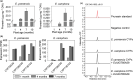
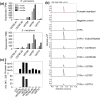
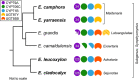
The economic and ecologically important genus Eucalyptus is rich in structurally diverse specialized metabolites. While some specialized metabolite classes are highly prevalent across the genus, the cyanogenic glucoside prunasin is only produced by c. 3% of species. To investigate the evolutionary mechanisms behind prunasin biosynthesis in Eucalyptus, we compared de novo assembled transcriptomes, together with online resources between cyanogenic and acyanogenic species. Identified genes were characterized in vivo and in vitro. Pathway characterization of cyanogenic Eucalyptus camphora and Eucalyptus yarraensis showed for the first time that the final glucosylation step from mandelonitrile to prunasin is catalyzed by a novel UDP-glucosyltransferase UGT87. This step is typically catalyzed by a member of the UGT85 family, including in Eucalyptus cladocalyx. The upstream conversion of phenylalanine to mandelonitrile is catalyzed by three cytochrome P450 (CYP) enzymes from the CYP79, CYP706, and CYP71 families, as previously shown. Analysis of acyanogenic Eucalyptus species revealed the loss of different ortholog prunasin biosynthetic genes. The recruitment of UGTs from different families for prunasin biosynthesis in Eucalyptus demonstrates important pathway heterogeneities and unprecedented dynamic pathway evolution of chemical defense within a single genus. Overall, this study provides relevant insights into the tremendous adaptability of these long-lived trees.
Keywords: Eucalyptus; UDP-glycosyltransferase; UGT87; chemical defense; cyanogenic glucoside; cytochrome P450; evolution; plant-specialized metabolism.
© 2022 The Authors. New Phytologist © 2022 New Phytologist Foundation.
Figures

References
-
- Bak S, Kahn RA, Nielsen HL, Møller BL, Halkier BA. 1998. Cloning of three A‐type cytochromes P450, CYP71E1, CYP98, and CYP99 from Sorghum bicolor (L.) Moench by a PCR approach and identification by expression in Escherichia coli of CYP71E1 as a multifunctional cytochrome P450 in the biosynthesis of the cyanogenic glucoside dhurrin. Plant Molecular Biology 36: 393–405. - PubMed
-
- Bak S, Olsen CE, Halkier BA, Møller BL. 2000. Transgenic tobacco and Arabidopsis plants expressing the two multifunctional sorghum cytochrome P450 enzymes, CYP79A1 and CYP71E1, are cyanogenic and accumulate metabolites derived from intermediates in dhurrin biosynthesis. Plant Physiology 123: 1437–1448. - PMC - PubMed
-
- Bennett AF. 2016. Eucalypts, wildlife and nature conservation: from individual trees to landscape patterns. Proceedings of the Royal Society of Victoria 128: 71–86.
-
- Bjarnholt N, Neilson EHJ, Crocoll C, Jørgensen K, Motawia MS, Olsen CE, Dixon DP, Edwards R, Møller BL. 2018. Glutathione transferases catalyze recycling of auto‐toxic cyanogenic glucosides in sorghum. The Plant Journal 94: 1109–1125. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

