Analysis of a hit-and-run tumor model by HPV in oropharyngeal cancers
- PMID: 36305515
- PMCID: PMC9828080
- DOI: 10.1002/jmv.28260
Analysis of a hit-and-run tumor model by HPV in oropharyngeal cancers
Abstract
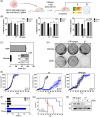
Several viruses are known to be associated with the development of certain cancers, including human papilloma virus (HPV), an established causative agent for a range of anogenital and head and neck cancers. However, the causality has been based on the presence of the virus, or its genetic material, in the sampled tumors. We have long wondered if viruses cause cancer via a "hit and run" mechanism such that they are no longer present in the resulting tumors. Here, we hypothesize that the absence of viral genes from the tumor does not necessarily exclude the viral aetiology. To test this, we used an HPV-driven oropharyngeal cancer (OPC) tumor model and CRISPR to delete the viral oncogene, E7. Indeed, the genetic removal of HPV E7 oncogene eliminates tumors in vivo. Remarkably, E7 deleted tumors recurred over time and develop new mutations not previously seen in HPV+ OPC tumors. Importantly, a number of these new mutations are found to be already present in HPV- OPC tumors.
Keywords: E7; human papilloma virus; oropharyngeal cancer.
© 2022 The Authors. Journal of Medical Virology published by Wiley Periodicals LLC.
Conflict of interest statement
NAJM receives payment from UniQuest Pty Ltd. and is also a consultant for Prorenata Ltd.
Figures

References
-
- de Martel C, Georges D, Bray F, Ferlay J, Clifford GM. Global burden of cancer attributable to infections in 2018: a worldwide incidence analysis. Lancet Glob Health. 2020;8(2):e180‐e190. - PubMed
-
- Ferreira DA, Tayyar Y, Idris A, McMillan NAJ. A “hit‐and‐run” affair—a possible link for cancer progression in virally driven cancers. Biochim Biophys Acta, Rev Cancer. 2021;1875(1):188476. - PubMed
-
- Galloway DA, McDougall JK. The oncogenic potential of herpes simplex viruses: evidence for a ‘hit‐and‐run' mechanism. Nature. 1983;302(5903):21‐24. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

