Intrinsic protein disorder uncouples affinity from binding specificity
- PMID: 36305763
- PMCID: PMC9601785
- DOI: 10.1002/pro.4455
Intrinsic protein disorder uncouples affinity from binding specificity
Abstract
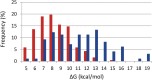
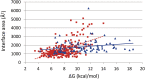
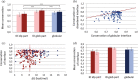
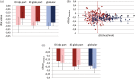
Intrinsically disordered proteins (IDPs) and intrinsically disordered regions (IDRs) of proteins often function by molecular recognition, in which they undergo induced folding. Based on prior generalizations, the idea prevails in the IDP field that due to the entropic penalty of induced folding, the major functional advantage associated with this binding mode is "uncoupling" specificity from binding strength. Nevertheless, both weaker binding and high specificity of IDPs/IDRs rest on limited experimental observations, making these assumptions more speculations than evidence-supported facts. The issue is also complicated by the rather vague concept of specificity that lacks an exact measure, such as the Kd for binding strength. We addressed these issues by creating and analyzing a comprehensive dataset of well-characterized ID/globular protein complexes, for which both the atomic structure of the complex and free energy (ΔG, Kd ) of interaction is known. Through this analysis, we provide evidence that the affinity distributions of IDP/globular and globular/globular complexes show different trends, whereas specificity does not connote to weaker binding strength of IDPs/IDRs. Furthermore, protein disorder extends the spectrum in the direction of very weak interactions, which may have important regulatory consequences and suggest that, in a biological sense, strict correlation of specificity and binding strength are uncoupled by structural disorder.
Keywords: IDPs; binding strength; conservation; disordered protein complexes; specificity; structural disorder.
© 2022 The Authors. Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Comment in
-
Reply to "Intrinsic protein disorder uncouples affinity from binding specificity".Protein Sci. 2023 Apr;32(4):e4601. doi: 10.1002/pro.4601. Protein Sci. 2023. PMID: 36808785 Free PMC article. No abstract available.
-
Comment on "Intrinsic protein disorder uncouples affinity from binding specificity".Protein Sci. 2023 Apr;32(4):e4602. doi: 10.1002/pro.4602. Protein Sci. 2023. PMID: 36840744 Free PMC article. No abstract available.
References
-
- Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27(10):527–533. - PubMed
-
- Tompa P. Unstructural biology coming of age. Curr Opin Struct Biol. 2011;21(3):419–425. - PubMed
-
- Chakrabarti P, Chakravarty D. Intrinsically disordered proteins/regions and insight into their biomolecular interactions. Biophys Chem. 2022;283:106769. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

