ATM-mediated ELL phosphorylation enhances its self-association through increased EAF1 interaction and inhibits global transcription during genotoxic stress
- PMID: 36305813
- PMCID: PMC9638944
- DOI: 10.1093/nar/gkac943
ATM-mediated ELL phosphorylation enhances its self-association through increased EAF1 interaction and inhibits global transcription during genotoxic stress
Erratum in
-
Correction to 'ATM-mediated ELL phosphorylation enhances its self-association through increased EAF1 interaction and inhibits global transcription during genotoxic stress'.Nucleic Acids Res. 2023 Jul 7;51(12):6507. doi: 10.1093/nar/gkad454. Nucleic Acids Res. 2023. PMID: 37194690 Free PMC article. No abstract available.
Abstract
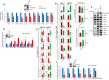
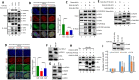
Mammalian cells immediately inhibit transcription upon exposure to genotoxic stress to avoid fatal collision between ongoing transcription and newly recruited DNA repair machineries to protect genomic integrity. However, mechanisms of this early transcriptional inhibition are poorly understood. In this study, we decipher a novel role of human EAF1, a positive regulator of ELL-dependent RNA Polymerase II-mediated transcription in vitro, in regulation of temporal inhibition of transcription during genotoxic stress. Our results show that, besides Super Elongation Complex (SEC) and Little Elongation Complex (LEC), human ELL (aka ELL1) also forms a complex with EAF1 alone. Interestingly, contrary to the in vitro studies, EAF1 inhibits ELL-dependent RNA polymerase II-mediated transcription of diverse target genes. Mechanistically, we show that intrinsic self-association property of ELL leads to its reduced interaction with other SEC components. EAF1 enhances ELL self-association and thus reduces its interaction with other SEC components leading to transcriptional inhibition. Physiologically, we show that upon exposure to genotoxic stress, ATM-mediated ELL phosphorylation-dependent enhanced EAF1 association results in reduced ELL interaction with other SEC components that lead to global transcriptional inhibition. Thus, we describe an important mechanism of dynamic transcriptional regulation during genotoxic stress involving post-translational modification of a key elongation factor.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





Similar articles
-
Negative Feedback Loop Mechanism between EAF1/2 and DBC1 in Regulating ELL Stability and Functions.Mol Cell Biol. 2022 Oct 20;42(10):e0015122. doi: 10.1128/mcb.00151-22. Epub 2022 Aug 29. Mol Cell Biol. 2022. PMID: 36036574 Free PMC article.
-
ELL-associated factors EAF1/2 negatively regulate HIV-1 transcription through inhibition of Super Elongation Complex formation.Biochim Biophys Acta Gene Regul Mech. 2020 May;1863(5):194508. doi: 10.1016/j.bbagrm.2020.194508. Epub 2020 Feb 19. Biochim Biophys Acta Gene Regul Mech. 2020. PMID: 32087315
-
Schizosaccharomyces pombe Pol II transcription elongation factor ELL functions as part of a rudimentary super elongation complex.Nucleic Acids Res. 2018 Nov 2;46(19):10095-10105. doi: 10.1093/nar/gky713. Nucleic Acids Res. 2018. PMID: 30102332 Free PMC article.
-
Collaboration of MLLT1/ENL, Polycomb and ATM for transcription and genome integrity.Nucleus. 2016 Apr 25;7(2):138-45. doi: 10.1080/19491034.2016.1177681. Nucleus. 2016. PMID: 27310306 Free PMC article. Review.
-
The super elongation complex (SEC) family in transcriptional control.Nat Rev Mol Cell Biol. 2012 Sep;13(9):543-7. doi: 10.1038/nrm3417. Epub 2012 Aug 16. Nat Rev Mol Cell Biol. 2012. PMID: 22895430 Review.
Cited by
-
Post-translational modification-dependent oligomerization switch in regulation of global transcription and DNA damage repair during genotoxic stress.Nat Commun. 2024 May 15;15(1):4128. doi: 10.1038/s41467-024-48530-8. Nat Commun. 2024. PMID: 38750015 Free PMC article.
-
Unveiling the role of GAS41 in cancer progression.Cancer Cell Int. 2023 Oct 18;23(1):245. doi: 10.1186/s12935-023-03098-z. Cancer Cell Int. 2023. PMID: 37853482 Free PMC article. Review.
-
Keep calm and reboot - how cells restart transcription after DNA damage and DNA repair.FEBS Lett. 2025 Jan;599(2):275-294. doi: 10.1002/1873-3468.14964. Epub 2024 Jul 11. FEBS Lett. 2025. PMID: 38991979 Free PMC article. Review.
References
-
- Svejstrup J.Q. The interface between transcription and mechanisms maintaining genome integrity. Trends Biochem. Sci. 2010; 35:333–338. - PubMed
-
- Wilson M.D., Harreman M., Svejstrup J.Q.. Ubiquitylation and degradation of elongating RNA polymerase II: the last resort. Biochim. Biophys. Acta. 2013; 1829:151–157. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

