A guard protein mediated quality control mechanism monitors 5'-capping of pre-mRNAs
- PMID: 36305816
- PMCID: PMC9638935
- DOI: 10.1093/nar/gkac952
A guard protein mediated quality control mechanism monitors 5'-capping of pre-mRNAs
Abstract
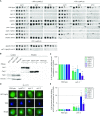
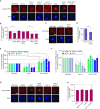
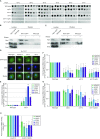
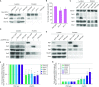
Efficient gene expression requires properly matured mRNAs for functional transcript translation. Several factors including the guard proteins monitor maturation and act as nuclear retention factors for unprocessed pre-mRNAs. Here we show that the guard protein Npl3 monitors 5'-capping. In its absence, uncapped transcripts resist degradation, because the Rat1-Rai1 5'-end degradation factors are not efficiently recruited to these faulty transcripts. Importantly, in npl3Δ, these improperly capped transcripts escape this quality control checkpoint and leak into the cytoplasm. Our data suggest a model in which Npl3 associates with the Rai1 bound pre-mRNAs. In case the transcript was properly capped and is thus CBC (cap binding complex) bound, Rai1 dissociates from Npl3 allowing the export factor Mex67 to interact with this guard protein and support nuclear export. In case Npl3 does not detect proper capping through CBC attachment, Rai1 binding persists and Rat1 can join this 5'-complex to degrade the faulty transcript.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Niño C.A., Hérissant L., Babour A., Dargemont C.. mRNA nuclear export in yeast. Chem. Rev. 2013; 113:8523–8545. - PubMed
-
- Tutucci E., Stutz F.. Keeping mRNPs in check during assembly and nuclear export. Nat. Rev. 2011; 12:377–384. - PubMed
-
- Zander G., Hackmann A., Bender L., Becker D., Lingner T., Salinas G., Krebber H.. mRNA quality control is bypassed for immediate export of stress-responsive transcripts. Nature. 2016; 540:593–596. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

