A Proximity biotinylation assay with a host protein bait reveals multiple factors modulating enterovirus replication
- PMID: 36306280
- PMCID: PMC9645661
- DOI: 10.1371/journal.ppat.1010906
A Proximity biotinylation assay with a host protein bait reveals multiple factors modulating enterovirus replication
Abstract
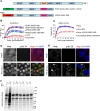
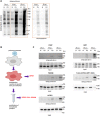
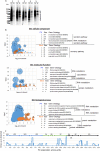
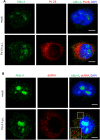
As ultimate parasites, viruses depend on host factors for every step of their life cycle. On the other hand, cells evolved multiple mechanisms of detecting and interfering with viral replication. Yet, our understanding of the complex ensembles of pro- and anti-viral factors is very limited in virtually every virus-cell system. Here we investigated the proteins recruited to the replication organelles of poliovirus, a representative of the genus Enterovirus of the Picornaviridae family. We took advantage of a strict dependence of enterovirus replication on a host protein GBF1, and established a stable cell line expressing a truncated GBF1 fused to APEX2 peroxidase that effectively supported viral replication upon inhibition of the endogenous GBF1. This construct biotinylated multiple host and viral proteins on the replication organelles. Among the viral proteins, the polyprotein cleavage intermediates were overrepresented, suggesting that the GBF1 environment is linked to viral polyprotein processing. The proteomics characterization of biotinylated host proteins identified multiple proteins previously associated with enterovirus replication, as well as more than 200 new factors recruited to the replication organelles. RNA metabolism proteins, many of which normally localize in the nucleus, constituted the largest group, underscoring the massive release of nuclear factors into the cytoplasm of infected cells and their involvement in viral replication. Functional analysis of several newly identified proteins revealed both pro- and anti-viral factors, including a novel component of infection-induced stress granules. Depletion of these proteins similarly affected the replication of diverse enteroviruses indicating broad conservation of the replication mechanisms. Thus, our data significantly expand the knowledge of the composition of enterovirus replication organelles, provide new insights into viral replication, and offer a novel resource for identifying targets for anti-viral interventions.
Copyright: © 2022 Moghimi et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
The development of resistance to an inhibitor of a cellular protein reveals a critical interaction between the enterovirus protein 2C and a small GTPase Arf1.PLoS Pathog. 2023 Sep 18;19(9):e1011673. doi: 10.1371/journal.ppat.1011673. eCollection 2023 Sep. PLoS Pathog. 2023. PMID: 37721955 Free PMC article.
-
Enterovirus Infection Induces Massive Recruitment of All Isoforms of Small Cellular Arf GTPases to the Replication Organelles.J Virol. 2020 Dec 22;95(2):e01629-20. doi: 10.1128/JVI.01629-20. Print 2020 Dec 22. J Virol. 2020. PMID: 33087467 Free PMC article.
-
Recruitment of PI4KIIIβ to coxsackievirus B3 replication organelles is independent of ACBD3, GBF1, and Arf1.J Virol. 2014 Mar;88(5):2725-36. doi: 10.1128/JVI.03650-13. Epub 2013 Dec 18. J Virol. 2014. PMID: 24352456 Free PMC article.
-
Combating enterovirus replication: state-of-the-art on antiviral research.Biochem Pharmacol. 2012 Jan 15;83(2):185-92. doi: 10.1016/j.bcp.2011.08.016. Epub 2011 Aug 26. Biochem Pharmacol. 2012. PMID: 21889497 Review.
-
The life cycle of non-polio enteroviruses and how to target it.Nat Rev Microbiol. 2018 Jun;16(6):368-381. doi: 10.1038/s41579-018-0005-4. Nat Rev Microbiol. 2018. PMID: 29626210 Review.
Cited by
-
Microbial Pathogenesis in the Era of Spatial Omics.Infect Immun. 2023 Jul 18;91(7):e0044222. doi: 10.1128/iai.00442-22. Epub 2023 May 31. Infect Immun. 2023. PMID: 37255461 Free PMC article. Review.
-
The development of resistance to an inhibitor of a cellular protein reveals a critical interaction between the enterovirus protein 2C and a small GTPase Arf1.PLoS Pathog. 2023 Sep 18;19(9):e1011673. doi: 10.1371/journal.ppat.1011673. eCollection 2023 Sep. PLoS Pathog. 2023. PMID: 37721955 Free PMC article.
-
Coxsackievirus B regulates host circAKAP10 expression to promote viral replication.Arch Virol. 2025 Jun 27;170(8):166. doi: 10.1007/s00705-025-06348-9. Arch Virol. 2025. PMID: 40576714 No abstract available.
-
Proteomic Analysis Revealed the Potential Role of MAGE-D2 in the Therapeutic Targeting of Triple-Negative Breast Cancer.Mol Cell Proteomics. 2024 Jan;23(1):100703. doi: 10.1016/j.mcpro.2023.100703. Epub 2023 Dec 20. Mol Cell Proteomics. 2024. PMID: 38128647 Free PMC article.
-
Stress granules attenuate protein nanoparticle induced osmotic imbalance via membrane potential restoration.Sci Rep. 2025 Aug 1;15(1):28125. doi: 10.1038/s41598-025-12903-w. Sci Rep. 2025. PMID: 40750804 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

