Deep generative molecular design reshapes drug discovery
- PMID: 36306797
- PMCID: PMC9797947
- DOI: 10.1016/j.xcrm.2022.100794
Deep generative molecular design reshapes drug discovery
Abstract
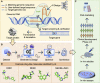
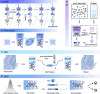
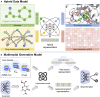
Recent advances and accomplishments of artificial intelligence (AI) and deep generative models have established their usefulness in medicinal applications, especially in drug discovery and development. To correctly apply AI, the developer and user face questions such as which protocols to consider, which factors to scrutinize, and how the deep generative models can integrate the relevant disciplines. This review summarizes classical and newly developed AI approaches, providing an updated and accessible guide to the broad computational drug discovery and development community. We introduce deep generative models from different standpoints and describe the theoretical frameworks for representing chemical and biological structures and their applications. We discuss the data and technical challenges and highlight future directions of multimodal deep generative models for accelerating drug discovery.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests E.F.F. has a CRADA arrangement with ChromaDex (USA) and is consultant to Aladdin Healthcare Technologies (UK and Germany), the Vancouver Dementia Prevention Centre (Canada), Intellectual Labs (Norway), and MindRank AI (China). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. S.K. and W.C. are employees of IBM TJ Watson Research Center. The other authors declare no competing interests.
Figures

References
-
- Avorn J. The $2.6 billion pill–methodologic and policy considerations. N. Engl. J. Med. 2015;372:1877–1879. - PubMed
-
- Fleming N. How artificial intelligence is changing drug discovery. Nature. 2018;557:S55–S57. - PubMed
-
- Hie B., Zhong E.D., Berger B., Bryson B. Learning the language of viral evolution and escape. Science. 2021;371:284–288. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

