Smoking modulates different secretory subpopulations expressing SARS-CoV-2 entry genes in the nasal and bronchial airways
- PMID: 36307504
- PMCID: PMC9615627
- DOI: 10.1038/s41598-022-17832-6
Smoking modulates different secretory subpopulations expressing SARS-CoV-2 entry genes in the nasal and bronchial airways
Abstract
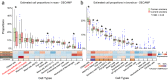
SARS-CoV-2 infection and disease severity are influenced by viral entry (VE) gene expression patterns in the airway epithelium. The similarities and differences of VE gene expression (ACE2, TMPRSS2, and CTSL) across nasal and bronchial compartments have not been fully characterized using matched samples from large cohorts. Gene expression data from 793 nasal and 1673 bronchial brushes obtained from individuals participating in lung cancer screening or diagnostic workup revealed that smoking status (current versus former) was the only clinical factor significantly and reproducibly associated with VE gene expression. The expression of ACE2 and TMPRSS2 was higher in smokers in the bronchus but not in the nose. scRNA-seq of nasal brushings indicated that ACE2 co-expressed genes were highly expressed in club and C15orf48+ secretory cells while TMPRSS2 co-expressed genes were highly expressed in keratinizing epithelial cells. In contrast, these ACE2 and TMPRSS2 modules were highly expressed in goblet cells in scRNA-seq from bronchial brushings. Cell-type deconvolution of the gene expression data confirmed that smoking increased the abundance of several secretory cell populations in the bronchus, but only goblet cells in the nose. The association of ACE2 and TMPRSS2 with smoking in the bronchus is due to their high expression in goblet cells which increase in abundance in current smoker airways. In contrast, in the nose, these genes are not predominantly expressed in cell populations modulated by smoking. In individuals with elevated lung cancer risk, smoking-induced VE gene expression changes in the nose likely have minimal impact on SARS-CoV-2 infection, but in the bronchus, smoking may lead to higher viral loads and more severe disease.
© 2022. The Author(s).
Conflict of interest statement
Avrum E. Spira is an employee of Johnson and Johnson Services, Inc, and has received personal fees from Veracyte Inc outside the submitted work. Marc E. Lenburg reports personal fees from Veracyte Inc outside the submitted work. Grant Duclos is now an employee of AstraZeneca. None declared (Ke Xu, Xingyi Shi, Christopher Husted, Rui Hong, Yichen Wang, Boting Ning, Travis Sullivan, Kimberly Rieger-Christ, Fenghai Duan, Helga Marques, Adam C. Gower, Xiaohui Xiao, Hanqiao Liu, Gang Liu, Michael Platt, Sarah Mazzilli, Ehab Billatos, Joshua D. Campbell, Jennifer E. Beane).
Figures



Update of
-
Smoking Modulates Different Secretory Subpopulations Expressing SARS-CoV-2 Entry Genes in the Nasal and Bronchial Airways.Res Sq [Preprint]. 2021 Oct 28:rs.3.rs-887718. doi: 10.21203/rs.3.rs-887718/v1. Res Sq. 2021. Update in: Sci Rep. 2022 Oct 28;12(1):18168. doi: 10.1038/s41598-022-17832-6. PMID: 34729557 Free PMC article. Updated. Preprint.
References
-
- Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., Schiergens, T. S., Herrler, G., Wu, N. H., Nitsche, A., Müller, M. A., Drosten, C., Pöhlmann, S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and Is blocked by a clinically proven protease inhibitor. Cell. 181(2), 271–280.e8 10.1016/j.cell.2020.02.052 (2020). - PMC - PubMed
-
- Jia HP, Look DC, Shi L, Hickey M, Pewe L, Netland J, Farzan M, Wohlford-Lenane C, Perlman S, McCray PB., Jr ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J. Virol. 2005;79(23):14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. - DOI - PMC - PubMed
-
- Smith, J. C., Sausville, E. L., Girish, V., Yuan, M. L., Vasudevan, A., John, K. M. & Sheltzer, J. M. Cigarette smoke exposure and inflammatory signaling increase the expression of the SARS-CoV-2 receptor ACE2 in the respiratory tract. Dev Cell. 53(5), 514–529.e.3 10.1016/j.devcel.2020.05.012 (2020). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

