SARS-CoV-2 variant evasion of monoclonal antibodies based on in vitro studies
- PMID: 36307535
- PMCID: PMC9616429
- DOI: 10.1038/s41579-022-00809-7
SARS-CoV-2 variant evasion of monoclonal antibodies based on in vitro studies
Abstract
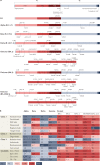
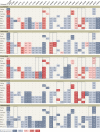
Monoclonal antibodies (mAbs) offer a treatment option for individuals with severe COVID-19 and are especially important in high-risk individuals where vaccination is not an option. Given the importance of understanding the evolution of resistance to mAbs by SARS-CoV-2, we reviewed the available in vitro neutralization data for mAbs against live variants and viral constructs containing spike mutations of interest. Unfortunately, evasion of mAb-induced protection is being reported with new SARS-CoV-2 variants. The magnitude of neutralization reduction varied greatly among mAb-variant pairs. For example, sotrovimab retained its neutralization capacity against Omicron BA.1 but showed reduced efficacy against BA.2, BA.4 and BA.5, and BA.2.12.1. At present, only bebtelovimab has been reported to retain its efficacy against all SARS-CoV-2 variants considered here. Resistance to mAb neutralization was dominated by the action of epitope single amino acid substitutions in the spike protein. Although not all observed epitope mutations result in increased mAb evasion, amino acid substitutions at non-epitope positions and combinations of mutations also contribute to evasion of neutralization. This Review highlights the implications for the rational design of viral genomic surveillance and factors to consider for the development of novel mAb therapies.
© 2022. Springer Nature Limited.
Conflict of interest statement
R.K.G. has received honoraria for educational activities from Moderna, GSK and Janssen. The other authors declare no competing interests.
Figures


References
-
- World Health Organization. WHO COVID-19 dashboard. World Health Organizationhttps://covid19.who.int/ (2022).
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

