Live slow-frozen human tumor tissues viable for 2D, 3D, ex vivo cultures and single-cell RNAseq
- PMID: 36307545
- PMCID: PMC9616892
- DOI: 10.1038/s42003-022-04025-0
Live slow-frozen human tumor tissues viable for 2D, 3D, ex vivo cultures and single-cell RNAseq
Abstract
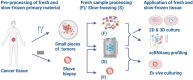
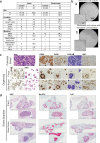
Biobanking of surplus human healthy and disease-derived tissues is essential for diagnostics and translational research. An enormous amount of formalin-fixed and paraffin-embedded (FFPE), Tissue-Tek OCT embedded or snap-frozen tissues are preserved in many biobanks worldwide and have been the basis of translational studies. However, their usage is limited to assays that do not require viable cells. The access to intact and viable human material is a prerequisite for translational validation of basic research, for novel therapeutic target discovery, and functional testing. Here we show that surplus tissues from multiple solid human cancers directly slow-frozen after resection can subsequently be used for different types of methods including the establishment of 2D, 3D, and ex vivo cultures as well as single-cell RNA sequencing with similar results when compared to freshly analyzed material.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
The impact of RNA extraction method on accurate RNA sequencing from formalin-fixed paraffin-embedded tissues.BMC Cancer. 2019 Dec 5;19(1):1189. doi: 10.1186/s12885-019-6363-0. BMC Cancer. 2019. PMID: 31805884 Free PMC article.
-
Fusion transcript discovery using RNA sequencing in formalin-fixed paraffin-embedded specimen.Crit Rev Oncol Hematol. 2021 Apr;160:103303. doi: 10.1016/j.critrevonc.2021.103303. Epub 2021 Mar 20. Crit Rev Oncol Hematol. 2021. PMID: 33757837 Review.
-
Cryopreservation of Viable Human Tissues: Renewable Resource for Viable Tissue, Cell Lines, and Organoid Development.Biopreserv Biobank. 2020 Jun;18(3):222-227. doi: 10.1089/bio.2019.0062. Epub 2020 Apr 17. Biopreserv Biobank. 2020. PMID: 32302515 Free PMC article.
-
Performance comparison of three DNA extraction kits on human whole-exome data from formalin-fixed paraffin-embedded normal and tumor samples.PLoS One. 2018 Apr 5;13(4):e0195471. doi: 10.1371/journal.pone.0195471. eCollection 2018. PLoS One. 2018. PMID: 29621323 Free PMC article.
-
Microtomy: Cutting Formalin-Fixed, Paraffin-Embedded Sections.Methods Mol Biol. 2019;1897:269-278. doi: 10.1007/978-1-4939-8935-5_23. Methods Mol Biol. 2019. PMID: 30539451 Review.
Cited by
-
DNA Quantity and Quality Comparisons between Cryopreserved and FFPE Tumors from Matched Pan-Cancer Samples.Curr Oncol. 2024 Apr 28;31(5):2441-2452. doi: 10.3390/curroncol31050183. Curr Oncol. 2024. PMID: 38785464 Free PMC article.
-
Impact of soft tissue homogenization methods on RNA quality.Mol Biol Rep. 2025 Apr 25;52(1):425. doi: 10.1007/s11033-025-10508-0. Mol Biol Rep. 2025. PMID: 40278925 Free PMC article.
-
Investigating the tumor-immune microenvironment through extracellular vesicles from frozen patient biopsies and 3D cultures.Front Immunol. 2023 May 25;14:1176175. doi: 10.3389/fimmu.2023.1176175. eCollection 2023. Front Immunol. 2023. PMID: 37304281 Free PMC article.
-
Dynamic thresholding and tissue dissociation optimization for CITE-seq identifies differential surface protein abundance in metastatic melanoma.Commun Biol. 2023 Aug 10;6(1):830. doi: 10.1038/s42003-023-05182-6. Commun Biol. 2023. PMID: 37563418 Free PMC article.
-
Tebentafusp elicits on-target cutaneous immune responses driven by cytotoxic T cells in uveal melanoma patients.J Clin Invest. 2025 Apr 29;135(12):e181464. doi: 10.1172/JCI181464. eCollection 2025 Jun 16. J Clin Invest. 2025. PMID: 40311102 Free PMC article. Clinical Trial.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

