Cloning and characterization of thermophilic endoglucanase and its application in the transformation of ginsenosides
- PMID: 36307574
- PMCID: PMC9617004
- DOI: 10.1186/s13568-022-01473-z
Cloning and characterization of thermophilic endoglucanase and its application in the transformation of ginsenosides
Abstract
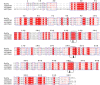
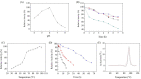
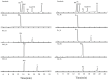
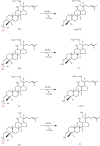
A novel endoglucanase (BcelFp) was identified from Fervidobaterium pennivorans DSM9078 which had biotransformation activity for protopanaxadiol (PPD)-type ginsenosides. Sequence analysis of BcelFp revealed that it could be classified into glycoside hydrolase family 5 (GH5). The gene encoding a 323-amino acid protein was cloned and expressed in Escherichia coli. The recombinant enzyme was purified, and its molecular weight was approximate 37 kDa. The recombinant BcelFp exhibited an optimal activity at 95 oC and pH 5.5 and showed high thermostability. The endoglucanase had high selectivity for cleaving the outer glucose moiety at the C3 carbon of ginsenoside Rb1, Rb2, Rc and Rd, which produced stronger pharmacologically active gypenoside XVII (GypXVII), Compound O (CO), Compound Mc1 (CMc1) and F2, respectively. The Km values for Rb1, Rb2, Rc and Rd were 3.66 ± 0.04 µM, 4.02 ± 0.12 µM, 5.95 ± 0.03 µM, 0.67 ± 0.006 µM, respectively. The kcat/Km value of BcelFp for ginsenoside Rd was 27.91 mM-1s-1, which was much higher than that of the previously enzymes. This study was the first report of the highly efficient and selective transformation of GypXVII, CO, CMc1 and F2 from Rb1, Rb2, Rc and Rd by a GH5-family thermophilic endoglucanase.
Keywords: Biotransformation; Endoglucanase; Fervidobaterium pennivorans DSM9078; Ginsenoside; Glycoside hydrolase family.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Cleland WW (1979) Statistical analysis of enzyme kinetic data. Methods Enzymol 63. 10.1002/9780470122747.ch1. :103 – 38 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

