SIAH proteins regulate the degradation and intra-mitochondrial aggregation of PINK1: Implications for mitochondrial pathology in Parkinson's disease
- PMID: 36307912
- PMCID: PMC9741505
- DOI: 10.1111/acel.13731
SIAH proteins regulate the degradation and intra-mitochondrial aggregation of PINK1: Implications for mitochondrial pathology in Parkinson's disease
Abstract
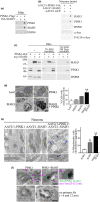
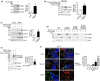
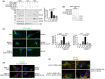
Parkinson's disease (PD) is characterized by degeneration of neurons, particularly dopaminergic neurons in the substantia nigra. PD brains show accumulation of α-synuclein in Lewy bodies and accumulation of dysfunctional mitochondria. However, the mechanisms leading to mitochondrial pathology in sporadic PD are poorly understood. PINK1 is a key for mitophagy activation and recycling of unfit mitochondria. The activation of mitophagy depends on the accumulation of uncleaved PINK1 at the outer mitochondrial membrane and activation of a cascade of protein ubiquitination at the surface of the organelle. We have now found that SIAH3, a member of the SIAH proteins but lacking ubiquitin-ligase activity, is increased in PD brains and cerebrospinal fluid and in neurons treated with α-synuclein preformed fibrils (α-SynPFF). We also observed that SIAH3 is aggregated together with PINK1 in the mitochondria of PD brains. SIAH3 directly interacts with PINK1, leading to their intra-mitochondrial aggregation in cells and neurons and triggering a cascade of toxicity with PINK1 inactivation along with mitochondrial depolarization and neuronal death. We also found that SIAH1 interacts with PINK1 and promotes ubiquitination and proteasomal degradation of PINK1. Similar to the dimerization of SIAH1/SIAH2, SIAH3 interacts with SIAH1, promoting its translocation to mitochondria and preventing its ubiquitin-ligase activity toward PINK1. Our results support the notion that the increase in SIAH3 and intra-mitochondrial aggregation of SIAH3-PINK1 may mediate α-synuclein pathology by promoting proteotoxicity and preventing the elimination of dysfunctional mitochondria. We consider it possible that PINK1 activity is decreased in sporadic PD, which impedes proper mitochondrial renewal in the disease.
Keywords: PINK1; Parkinson's disease; SIAH1; SIAH3; intra-mitochondrial protein aggregation; mitochondrial dysfunction; mitophagy; ubiquitin.
© 2022 The Authors. Aging Cell published by Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
None declared.
Figures



References
-
- Di Maio, R. , Barrett, P. J. , Hoffman, E. K. , Barrett, C. W. , Zharikov, A. , Borah, A. , Hu, X. , McCoy, J. , Chu, C. T. , Burton, E. A. , Hastings, T. G. , & Greenamyre, J. T. (2016). Alpha‐synuclein binds to TOM20 and inhibits mitochondrial protein import in Parkinson's disease. Science Translational Medicine, 8(342), 342ra378. 10.1126/scitranslmed.aaf3634 - DOI - PMC - PubMed
-
- Gandhi, S. , Muqit, M. M. , Stanyer, L. , Healy, D. G. , Abou‐Sleiman, P. M. , Hargreaves, I. , Heales, S. , Ganguly, M. , Parsons, L. , Lees, A. J. , Latchman, D. S. , Holton, J. L. , Wood, N. W. , & Revesz, T. (2006). PINK1 protein in normal human brain and Parkinson's disease. Brain, 129(Pt 7), 1720–1731. 10.1093/brain/awl114 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

