Global Phosphoproteomics Unveils Kinase-Regulated Networks in Systemic Lupus Erythematosus
- PMID: 36309313
- PMCID: PMC9712766
- DOI: 10.1016/j.mcpro.2022.100434
Global Phosphoproteomics Unveils Kinase-Regulated Networks in Systemic Lupus Erythematosus
Abstract
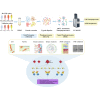
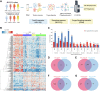
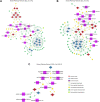
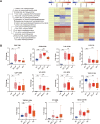
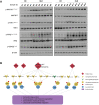
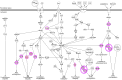
Systemic lupus erythematosus (SLE) is an autoimmune disorder characterized by immune complex deposition in multiple organs. Despite the severe symptoms caused by it, the underlying mechanisms of SLE, especially phosphorylation-dependent regulatory networks remain elusive. Herein, by combining high-throughput phosphoproteomics with bioinformatics approaches, we established the global phosphoproteome landscape of the peripheral blood mononuclear cells from a large number of SLE patients, including the remission stage (SLE_S), active stage (SLE_A), rheumatoid arthritis, and healthy controls, and thus a deep mechanistic insight into SLE signaling mechanism was yielded. Phosphorylation upregulation was preferentially in patients with SLE (SLE_S and SLE_A) compared with healthy controls and rheumatoid arthritis populations, resulting in an atypical enrichment in cell adhesion and migration signatures. Several specifically upregulated phosphosites were identified, and the leukocyte transendothelial migration pathway was enriched in the SLE_A group by expression pattern clustering analysis. Phosphosites identified by 4D-label-free quantification unveiled key kinases and kinase-regulated networks in SLE, then further validated by parallel reaction monitoring. Some of these validated phosphosites including vinculin S275, vinculin S579 and transforming growth factor beta-1-induced transcript 1 S68, primarily were phosphorylation of Actin Cytoskeleton -related proteins. Some predicted kinases including MAP3K7, TBK1, IKKβ, and GSK3β, were validated by Western blot using kinases phosphorylation sites-specific antibodies. Taken together, the study has yielded fundamental insights into the phosphosites, kinases, and kinase-regulated networks in SLE. The map of the global phosphoproteomics enables further understanding of this disease and will provide great help for seeking more potential therapeutic targets for SLE.
Keywords: SLE active stage; SLE remission stage; kinases; parallel reaction monitoring; phosphoproteomics.
Copyright © 2022. Published by Elsevier Inc.
Conflict of interest statement
Conflict of interest All authors declare no competing interests.
Figures





References
-
- Durca’ L., O'Dwyer T., Petri M. Management strategies and future directions for systemic lupus erythematosus in adults. Lancet. 2019;393:2332–2343. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

