Genomic evolution of SARS-CoV-2 in Reunion Island
- PMID: 36309317
- PMCID: PMC9598258
- DOI: 10.1016/j.meegid.2022.105381
Genomic evolution of SARS-CoV-2 in Reunion Island
Abstract
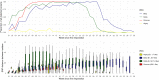
Island communities are interesting study sites for microbial evolution during epidemics, as their insular nature reduces the complexity of the population's connectivity. This was particularly true on Reunion Island during the first half of 2021, when international travel was restricted in order to mitigate the risk for SARS-CoV-2 introductions. Concurrently, the SARS-CoV-2 Beta variant became dominant and started to circulate at high levels for several months before being completely replaced by the Delta variant as of October 2021. Here, we explore some of the particularities of SARS-CoV-2 genomic evolution within the insular context of Reunion Island. We show that island isolation allowed the amplification and expansion of unique genetic lineages that remained uncommon across the globe. Islands are therefore potential hotspots for the emergence of new genetic variants, meaning that they will play a key role in the continued evolution and propagation of COVID-19 as the pandemic persists.
Keywords: Evolution; Genomics; Reunion Island; SARS-CoV-2.
Copyright © 2022 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare no conflict of interest.
Figures




References
-
- Aksamentov I., Roemer C., Hodcroft E.B., Neher R.A. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J. Open Source Softw. 2021;6:3773. doi: 10.21105/JOSS.03773. - DOI
-
- Althaus C.L., Baggio S., Reichmuth M.L., Hodcroft E.B., Riou J., Neher R.A., et al. A tale of two variants: spread of SARS-CoV-2 variants alpha in Geneva, Switzerland, and Beta in South Africa. medRxiv. 2021 doi: 10.1101/2021.06.10.21258468. 2021.06.10.21258468. - DOI
-
- Ciuffreda L., González-Montelongo R., Alcoba-Florez J., García-Martínez de Artola D., Gil-Campesino H., Rodríguez-Pérez H., et al. Tracing the trajectories of SARS-CoV-2 variants of concern between December 2020 and September 2021 in the Canary Islands (Spain) Front. Cell. Infect. Microbiol. 2022;12 doi: 10.3389/fcimb.2022.919346. - DOI - PMC - PubMed
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

