Remodeling of the osteoimmune microenvironment after biomaterials implantation in murine tibia: Single-cell transcriptome analysis
- PMID: 36311047
- PMCID: PMC9588995
- DOI: 10.1016/j.bioactmat.2022.10.009
Remodeling of the osteoimmune microenvironment after biomaterials implantation in murine tibia: Single-cell transcriptome analysis
Abstract
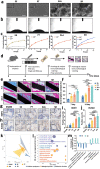
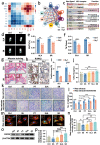
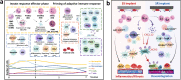
Osseointegration seems to be a foreign body reaction equilibrium due to the complicated interactions between the immune and skeletal systems. The heterogeneity of the osteoimmune microenvironment in the osseointegration of implant materials remains elusive. Here, a single-cell study involving 40043 cells is conducted, and a total of 10 distinct cell clusters are identified from five different groups. A preliminary description of the osteoimmune microenvironment revealed the diverse cellular heterogeneity and dynamic changes modulated by implant properties. The increased immature neutrophils, Ly6C + CCR2hi monocytes, and S100a8hi macrophages induce an aggressive inflammatory response and eventually lead to the formation of fibrous capsule around the stainless steel implant. The enrichment of mature neutrophils, FcgR1hi and differentiated immunomodulatory macrophages around the titanium implant indicates favorable osseointegration under moderate immune response. Neutrophil-depletion mice are conducted to explore the role of neutrophils in osseointegration. Neutrophils may improve bone formation by enhancing the recruitment of BMSCs via the CXCL12/CXCR3 signal axis. These findings contribute to a better knowledge of osteoimmunology and are valuable for the design and modification of 'osteoimmune-smart' biomaterials in the bone regeneration field.
Keywords: BMP2, Bone Morphogenetic Proteins 2; CXCL12, Chemokine (C-X-C mode) Ligand 12; CXCR, CXC Chemokine Receptor; FcgR, Fc Gamma Receptor; IFN-γ, Interferon-gamma; IL-1β, Interleukin-1 beta; Implant; MHC, Major Histocompatibility Complex; MIP, Macrophage inflammatory cytokines; MPO, Myeloperoxidase; NE, Neutrophil Elastase; NF-κB, Nuclear Factor Kappa-light-chain-enhancer of Activated B cells; NOD, Nucleotide Binding Oligomerization Domain; Neutrophil; OPG, Osteoprotegerin; Osseointegration; Osteoimmunology; RANKL, Nuclear Factor B receptor Activator Ligand; RUNX2, Runt-related Transcription Factor 2; S100a8, S100 Calcium Binding Protein A8; SDF-1α, Stromal Cell-derived Factor-1 alpha; STAT, Signal Transduction and Transcription Activator; Single-cell transcriptomics; TLR, Toll Like Receptor; TNFα, Tumor Necrosis Factor-alpha; TRAP, Tartrate Resistant Acid Phosphatase.
© 2022 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures






References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

