Alternative splicing reprogramming in fungal pathogen Sclerotinia sclerotiorum at different infection stages on Brassica napus
- PMID: 36311105
- PMCID: PMC9597501
- DOI: 10.3389/fpls.2022.1008665
Alternative splicing reprogramming in fungal pathogen Sclerotinia sclerotiorum at different infection stages on Brassica napus
Abstract
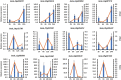
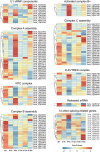
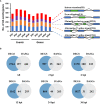
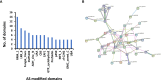
Alternative splicing (AS) is an important post-transcriptional mechanism promoting the diversity of transcripts and proteins to regulate various life processes in eukaryotes. Sclerotinia stem rot is a major disease of Brassica napus caused by Sclerotinia sclerotiorum, which causes severe yield loss in B. napus production worldwide. Although many transcriptome studies have been carried out on the growth, development, and infection of S. sclerotiorum, the genome-wide AS events of S. sclerotiorum remain poorly understood, particularly at the infection stage. In this study, transcriptome sequencing was performed to systematically explore the genome-scale AS events of S. sclerotiorum at five important infection stages on a susceptible oilseed rape cultivar. A total of 130 genes were predicted to be involved in AS from the S. sclerotiorum genome, among which 98 genes were differentially expressed and may be responsible for AS reprogramming for its successful infection. In addition, 641 differential alternative splicing genes (DASGs) were identified during S. sclerotiorum infection, accounting for 5.76% of all annotated S. sclerotiorum genes, and 71 DASGs were commonly found at all the five infection stages. The most dominant AS type of S. sclerotiorum was found to be retained introns or alternative 3' splice sites. Furthermore, the resultant AS isoforms of 21 DASGs became pseudogenes, and 60 DASGs encoded different putative proteins with different domains. More importantly, 16 DASGs of S. sclerotiorum were found to have signal peptides and possibly encode putative effectors to facilitate the infection of S. sclerotiorum. Finally, about 69.27% of DASGs were found to be non-differentially expressed genes, indicating that AS serves as another important way to regulate the infection of S. sclerotiorum on plants besides the gene expression level. Taken together, this study provides a genome-wide landscape for the AS of S. sclerotiorum during infection as well as an important resource for further elucidating the pathogenic mechanisms of S. sclerotiorum.
Keywords: Sclerotinia sclerotiorum; alternative splicing; reprogramming; secreted proteins; transcriptome sequencing.
Copyright © 2022 Cheng, Zhao, Gao, Zeng, Xu, Liu, Huang, Liu, Liu and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Screening of microRNAs and target genes involved in Sclerotinia sclerotiorum (Lib.) infection in Brassica napus L.BMC Plant Biol. 2023 Oct 9;23(1):479. doi: 10.1186/s12870-023-04501-7. BMC Plant Biol. 2023. PMID: 37807039 Free PMC article.
-
Integrated mRNA, sRNA, and degradome sequencing reveal oilseed rape complex responses to Sclerotinia sclerotiorum (Lib.) infection.Sci Rep. 2018 Jul 20;8(1):10987. doi: 10.1038/s41598-018-29365-y. Sci Rep. 2018. PMID: 30030454 Free PMC article.
-
Differential Alternative Splicing Genes and Isoform Regulation Networks of Rapeseed (Brassica napus L.) Infected with Sclerotinia sclerotiorum.Genes (Basel). 2020 Jul 13;11(7):784. doi: 10.3390/genes11070784. Genes (Basel). 2020. PMID: 32668742 Free PMC article.
-
Genetic breakthroughs in the Brassica napus-Sclerotinia sclerotiorum interactions.Front Plant Sci. 2023 Nov 23;14:1276055. doi: 10.3389/fpls.2023.1276055. eCollection 2023. Front Plant Sci. 2023. PMID: 38078117 Free PMC article. Review.
-
Sclerotinia Stem Rot Resistance in Rapeseed: Recent Progress and Future Prospects.J Agric Food Chem. 2021 Mar 17;69(10):2965-2978. doi: 10.1021/acs.jafc.0c07351. Epub 2021 Mar 5. J Agric Food Chem. 2021. PMID: 33667087 Review.
Cited by
-
Comprehensive mapping of Arabidopsis alternative splicing landscape reveals key insights into plant development and immunity.Plant Genome. 2025 Jun;18(2):e70022. doi: 10.1002/tpg2.70022. Plant Genome. 2025. PMID: 40156198 Free PMC article.
-
Genome-Wide Identification of Alternative Splicing in Botrytis cinerea During Infection Stage of Solanum lycopersicum.Microorganisms. 2025 Feb 7;13(2):360. doi: 10.3390/microorganisms13020360. Microorganisms. 2025. PMID: 40005727 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

