Genetic identification of SNP markers and candidate genes associated with sugarcane smut resistance using BSR-Seq
- PMID: 36311133
- PMCID: PMC9608552
- DOI: 10.3389/fpls.2022.1035266
Genetic identification of SNP markers and candidate genes associated with sugarcane smut resistance using BSR-Seq
Abstract
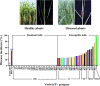
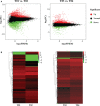
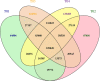
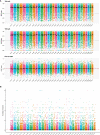
Sugarcane smut caused by Sporisorium scitamineum is one of the most severe fungal diseases worldwide. In this study, a cross was made between a smut-resistant variety YT93-159 and a smut-susceptible variety ROC22, and 312 progenies were obtained. Two bulks of progenies were then constructed, one consisted of 27 highly smut resistant progenies and the other 24 smut susceptible progenies. Total RNAs of the progenies of each bulk, were pooled and subject to bulked segregant RNA-sequence analysis (BSR-Seq). A total of 164.44 Gb clean data containing 2,341,449 SNPs and 64,999 genes were obtained, 7,295 of which were differentially expressed genes (DEGs). These DEGs were mainly enriched in stress-related metabolic pathways, including carbon metabolism, phenylalanine metabolism, plant hormone signal transduction, glutathione metabolism, and plant-pathogen interactions. Besides, 45,946 high-quality, credible SNPs, a 1.27 Mb region at Saccharum spontaneum chromosome Chr5B (68,904,827 to 70,172,982), and 129 candidate genes were identified to be associated with smut resistance. Among them, twenty-four genes, either encoding key enzymes involved in signaling pathways or being transcription factors, were found to be very closely associated with stress resistance. RT-qPCR analysis demonstrated that they played a positive role in smut resistance. Finally, a potential molecular mechanism of sugarcane and S. scitamineum interaction is depicted that activations of MAPK cascade signaling, ROS signaling, Ca2+ signaling, and PAL metabolic pathway and initiation of the glyoxalase system jointly promote the resistance to S. scitamineum in sugarcane. This study provides potential SNP markers and candidate gene resources for smut resistance breeding in sugarcane.
Keywords: BSR-seq; SNPs; expression pattern; key genes; molecular mechanism; smut resistance; sugarcane.
Copyright © 2022 Wu, Su, Pan, Xu, Zou, Que, Lin, Sun, Grisham, Xu and Que.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


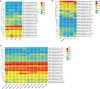
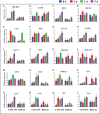

References
-
- Akalach M., Touil B. (1996). Occurrence and spread of sugarcane smut caused by Ustilago scitaminea in Morocco. Plant Dis. 80, 1363–1366. doi: 10.1094/PD-80-1363 - DOI
-
- Álvarez Viveros M. F., Inostroza-Blancheteau C., Timmermann T., González M., Arce-Johnson P. (2013). Overexpression of GlyI and GlyII genes in transgenic tomato (Solanum lycopersicum mill.) plants confers salt tolerance by decreasing oxidative stress. Mol. Biol. Rep. 40, 3281–3290. doi: 10.1007/s11033-012-2403-4 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

