Evolution of surrogate light chain in tetrapods and the relationship between lengths of CDR H3 and VpreB tails
- PMID: 36311706
- PMCID: PMC9614664
- DOI: 10.3389/fimmu.2022.1001134
Evolution of surrogate light chain in tetrapods and the relationship between lengths of CDR H3 and VpreB tails
Abstract
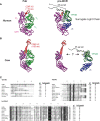
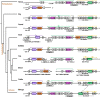
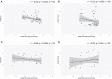
In the mammalian immune system, the surrogate light chain (SLC) shapes the antibody repertoire during B cell development by serving as a checkpoint for production of functional heavy chains (HC). Structural studies indicate that tail regions of VpreB contact and cover the third complementarity-determining region of the HC (CDR H3). However, some species, particularly bovines, have CDR H3 regions that may not be compatible with this HC-SLC interaction model. With immense structural and genetic diversity in antibody repertoires across species, we evaluated the genetic origins and sequence features of surrogate light chain components. We examined tetrapod genomes for evidence of conserved gene synteny to determine the evolutionary origin of VpreB1, VpreB2, and IGLL1, as well as VpreB3 and pre-T cell receptor alpha (PTCRA) genes. We found the genes for the SLC components (VpreB1, VpreB2, and IGLL1) only in eutherian mammals. However, genes for PTCRA occurred in all amniote groups and genes for VpreB3 occurred in all tetrapod groups, and these genes were highly conserved. Additionally, we found evidence of a new VpreB gene in non-mammalian tetrapods that is similar to the VpreB2 gene of eutherian mammals, suggesting VpreB2 may have appeared earlier in tetrapod evolution and may be a precursor to traditional VpreB2 genes in higher vertebrates. Among eutherian mammals, sequence conservation between VpreB1 and VpreB2 was low for all groups except rabbits and rodents, where VpreB2 was nearly identical to VpreB1 and did not share conserved synteny with VpreB2 of other species. VpreB2 of rabbits and rodents likely represents a duplicated variant of VpreB1 and is distinct from the VpreB2 of other mammals. Thus, rabbits and rodents have two variants of VpreB1 (VpreB1-1 and VpreB1-2) but no VpreB2. Sequence analysis of VpreB tail regions indicated differences in sequence content, charge, and length; where repertoire data was available, we observed a significant relationship between VpreB2 tail length and maximum DH length. We posit that SLC components co-evolved with immunoglobulin HC to accommodate the repertoire - particularly CDR H3 length and structure, and perhaps highly unusual HC (like ultralong HC of cattle) may bypass this developmental checkpoint altogether.
Keywords: Lambda5; VpreB; evolution; pre-B cell receptor; pre-T cell receptor alpha; surrogate light chain.
Copyright © 2022 Ott, Haakenson, Kelly, Christian, Criscitiello and Smider.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

