Increased prevalence of clonal hematopoiesis of indeterminate potential in hospitalized patients with COVID-19
- PMID: 36311800
- PMCID: PMC9614713
- DOI: 10.3389/fimmu.2022.968778
Increased prevalence of clonal hematopoiesis of indeterminate potential in hospitalized patients with COVID-19
Abstract
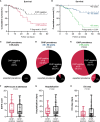
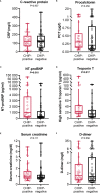
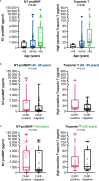
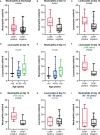
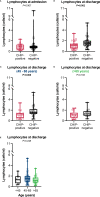
Clonal hematopoiesis of indeterminate potential (CHIP) leads to higher mortality, carries a cardiovascular risk and alters inflammation. All three aspects harbor overlaps with the clinical manifestation of COVID-19. This study aimed to identify the impact of CHIP on COVID-19 pathophysiology. 90 hospitalized patients were analyzed for CHIP. In addition, their disease course and outcome were evaluated. With a prevalence of 37.8%, the frequency of a CHIP-driver mutation was significantly higher than the prevalence expected based on median age (17%). CHIP increases the risk of hospitalization in the course of the disease but has no age-independent impact on the outcome within the group of hospitalized patients. Especially in younger patients (45 - 65 years), CHIP was associated with persistent lymphopenia. In older patients (> 65 years), on the other hand, CHIP-positive patients developed neutrophilia in the long run. To what extent increased values of cardiac biomarkers are caused by CHIP independent of age could not be elaborated solely based on this study. In conclusion, our results indicate an increased susceptibility to a severe course of COVID-19 requiring hospitalization associated with CHIP. Secondly, they link it to a differentially regulated cellular immune response under the pressure of SARS-CoV-2 infection. Hence, a patient's CHIP-status bears the potential to serve as biomarker for risk stratification and to early guide treatment of COVID-19 patients.
Keywords: CHIP; SARS-CoV-2; cardiac function; critically ill; immune system.
Copyright © 2022 Schenz, Rump, Siegler, Hemmerling, Rahmel, Thon, Nowak, Fischer, Hafner, Tichy, Bomans, Meggendorfer, Koos, von Groote, Zarbock, Fiedler, Zemva, Larmann, Merle, Adamzik, Müller-Tidow, Haferlach, Leuschner and Weigand.
Conflict of interest statement
Author TH declares part ownership of MLL Munich Leukemia Laboratory. Author MM is employed by MLL. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

