Uterine lavage identifies cancer mutations and increased TP53 somatic mutation burden in individuals with ovarian cancer
- PMID: 36311816
- PMCID: PMC9615025
- DOI: 10.1158/2767-9764.crc-22-0314
Uterine lavage identifies cancer mutations and increased TP53 somatic mutation burden in individuals with ovarian cancer
Abstract
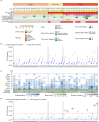
Current screening methods for ovarian cancer (OC) have failed to demonstrate a significant reduction in mortality. Uterine lavage combined with TP53 ultra-deep sequencing for the detection of disseminated OC cells has emerged as a promising tool, but this approach has not been tested for early-stage disease or non-serous histologies. In addition, lavages carry multiple background mutations, the significance of which is poorly understood. Uterine lavage was collected preoperatively in 34 patients undergoing surgery for suspected ovarian malignancy including 14 patients with benign disease and 20 patients with OC (6 non-serous and 14 high grade serous-like (serous)). Ultra-deep duplex sequencing (~3000x) with a panel of common OC genes identified the tumor mutation in 33% of non-serous (all early stage) and in 79% of serous cancers (including four early stage). In addition, all lavages carried multiple somatic mutations (average of 25 mutations per lavage), more than half of which corresponded to common cancer driver mutations. Driver mutations in KRAS, PIK3CA, PTEN, PPP2R1A and ARID1A presented as larger clones than non-driver mutations and with similar frequency in lavages from patients with and without OC, indicating prevalent somatic evolution in all patients. Driver TP53 mutations, however, presented as significantly larger clones and with higher frequency in lavages from individuals with OC, suggesting that TP53-specific clonal expansions are linked to ovarian cancer development. Our results demonstrate that lavages capture cancer cells, even from early-stage cancers, as well as other clonal expansions and support further exploration of TP53 mutation burden as a potential OC risk factor.
Keywords: Uterine lavage; clonal expansions; ovarian cancer; somatic evolution.
Conflict of interest statement
H.J. Gray reports other from UpToDate outside the submitted work. J.B. Liao reports grants from Merck, Precigen, AstraZeneca, Forty-Seven, Sumitomo Dainippon Pharma Oncology, Laekna Therapeutics, Instil Bio, Harpoon Therapeutics, and Genentech outside the submitted work. P. Speiser reports a patent to Three Lumen Balloon Catheter Apparatus US2015305725 issued and a patent to Non-Invasive Cancer Diagnosis EP51899 20130131 issued; and is shareholder of OVARTEC GmbH. E.M. Swisher reports grants from NIH during the conduct of the study. R.A. Risques reports grants from NCI, Rivkin Center, Minnesota Ovarian Cancer Alliance; and personal fees and other from TwinStrand Biosciences during the conduct of the study; grants from NCI and Rivkin Center; other from NanoString Technologies; and personal fees, non-financial support, and other from TwinStrand Biosciences outside the submitted work; in addition, R.A. Risques has a patent to WO/2018/175997 pending, licensed, and with royalties paid. No disclosures were reported by the other authors.
Figures



References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7–30. - PubMed
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. . Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. - PubMed
-
- Ovary SEER 5-Year Relative Survival Rates, 2012–2018. Surveillance, Epidemiology, and End Results (SEER) Program Populations (1969–2020) (www.seer.cancer.gov/popdata), National Cancer Institute, DCCPS, Surveillance Research Program; 2022.
-
- Buys SS, Partridge E, Black A, Johnson CC, Lamerato L, Isaacs C, et al. . Effect of screening on ovarian cancer mortality: the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Randomized Controlled Trial. JAMA 2011;305:2295–303. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

