Microfluidics for long-term single-cell time-lapse microscopy: Advances and applications
- PMID: 36312536
- PMCID: PMC9597311
- DOI: 10.3389/fbioe.2022.968342
Microfluidics for long-term single-cell time-lapse microscopy: Advances and applications
Abstract
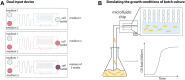
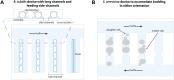
Cells are inherently dynamic, whether they are responding to environmental conditions or simply at equilibrium, with biomolecules constantly being made and destroyed. Due to their small volumes, the chemical reactions inside cells are stochastic, such that genetically identical cells display heterogeneous behaviors and gene expression profiles. Studying these dynamic processes is challenging, but the development of microfluidic methods enabling the tracking of individual prokaryotic cells with microscopy over long time periods under controlled growth conditions has led to many discoveries. This review focuses on the recent developments of one such microfluidic device nicknamed the mother machine. We overview the original device design, experimental setup, and challenges associated with this platform. We then describe recent methods for analyzing experiments using automated image segmentation and tracking. We further discuss modifications to the experimental setup that allow for time-varying environmental control, replicating batch culture conditions, cell screening based on their dynamic behaviors, and to accommodate a variety of microbial species. Finally, this review highlights the discoveries enabled by this technology in diverse fields, such as cell-size control, genetic mutations, cellular aging, and synthetic biology.
Keywords: cell screening; cellular dynamics; microfluidics; phenotypic heterogeneity; single-cell analysis; time-lapse microscopy.
Copyright © 2022 Allard, Papazotos and Potvin-Trottier.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Amir A. (2014). Cell size regulation in bacteria. Phys. Rev. Lett. 112, 208102. 10.1103/physrevlett.112.208102 - DOI
Publication types
LinkOut - more resources
Full Text Sources

