Mirubactin C rescues the lethal effect of cell wall biosynthesis mutations in Bacillus subtilis
- PMID: 36312962
- PMCID: PMC9609785
- DOI: 10.3389/fmicb.2022.1004737
Mirubactin C rescues the lethal effect of cell wall biosynthesis mutations in Bacillus subtilis
Abstract
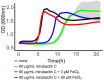
Growth of most rod-shaped bacteria is accompanied by the insertion of new peptidoglycan into the cylindrical cell wall. This insertion, which helps maintain and determine the shape of the cell, is guided by a protein machine called the rod complex or elongasome. Although most of the proteins in this complex are essential under normal growth conditions, cell viability can be rescued, for reasons that are not understood, by the presence of a high (mM) Mg2+ concentration. We screened for natural product compounds that could rescue the growth of mutants affected in rod-complex function. By screening > 2,000 extracts from a diverse collection of actinobacteria, we identified a compound, mirubactin C, related to the known iron siderophore mirubactin A, which rescued growth in the low micromolar range, and this activity was confirmed using synthetic mirubactin C. The compound also displayed toxicity at higher concentrations, and this effect appears related to iron homeostasis. However, several lines of evidence suggest that the mirubactin C rescuing activity is not due simply to iron sequestration. The results support an emerging view that the functions of bacterial siderophores extend well beyond simply iron binding and uptake.
Keywords: B. subtilis; cell wall mutants; chemical biological activity; mirubactin; siderophore.
Copyright © 2022 Kepplinger, Wen, Tyler, Kim, Brown, Banks, Dashti, Mackenzie, Wills, Kawai, Waldron, Allenby, Wu, Hall and Errington.
Conflict of interest statement
NA and B-YK were an employee of and JE scientific founder of and shareholder in Demuris Ltd. (now Odyssey Therapeutics Inc.). The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Bhavsar A. P., Beveridge J. T., Brown D. E. (2001). Precise deletion of tagD and controlled depletion of its product, glycerol 3-phosphate cytidylyltransferase, leads to irregular morphology and lysis of Bacillus subtilis grown at physiological temperature. J. Bacteriol. 183 6688–6693. 10.1128/JB.183.22.6688-6693.2001 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

