Thermal proteome profiling reveals Haemonchus orphan protein HCO_011565 as a target of the nematocidal small molecule UMW-868
- PMID: 36313370
- PMCID: PMC9616048
- DOI: 10.3389/fphar.2022.1014804
Thermal proteome profiling reveals Haemonchus orphan protein HCO_011565 as a target of the nematocidal small molecule UMW-868
Abstract
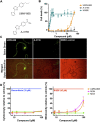
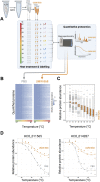
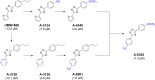
Parasitic roundworms (nematodes) cause destructive diseases, and immense suffering in humans and other animals around the world. The control of these parasites relies heavily on anthelmintic therapy, but treatment failures and resistance to these drugs are widespread. As efforts to develop vaccines against parasitic nematodes have been largely unsuccessful, there is an increased focus on discovering new anthelmintic entities to combat drug resistant worms. Here, we employed thermal proteome profiling (TPP) to explore hit pharmacology and to support optimisation of a hit compound (UMW-868), identified in a high-throughput whole-worm, phenotypic screen. Using advanced structural prediction and docking tools, we inferred an entirely novel, parasite-specific target (HCO_011565) of this anthelmintic small molecule in the highly pathogenic, blood-feeding barber's pole worm, and in other socioeconomically important parasitic nematodes. The "hit-to-target" workflow constructed here provides a unique prospect of accelerating the simultaneous discovery of novel anthelmintics and associated parasite-specific targets.
Keywords: anthelmintic discovery; in silico docking; structure modelling; target identification; thermal proteome profiling.
Copyright © 2022 Taki, Wang, Nguyen, Ang, Leeming, Nie, Byrne, Young, Zheng, Ma, Korhonen, Koehler, Williamson, Hofmann, Chang, Häberli, Keiser, Jabbar, Sleebs and Gasser.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Prediction and Prioritisation of Novel Anthelmintic Candidates from Public Databases Using Deep Learning and Available Bioactivity Data Sets.Int J Mol Sci. 2025 Mar 28;26(7):3134. doi: 10.3390/ijms26073134. Int J Mol Sci. 2025. PMID: 40243899 Free PMC article.
-
Comparative structure activity and target exploration of 1,2-diphenylethynes in Haemonchus contortus and Caenorhabditis elegans.Int J Parasitol Drugs Drug Resist. 2024 Aug;25:100534. doi: 10.1016/j.ijpddr.2024.100534. Epub 2024 Mar 19. Int J Parasitol Drugs Drug Resist. 2024. PMID: 38554597 Free PMC article.
-
A perspective on the discovery of selected compounds with anthelmintic activity against the barber's pole worm-Where to from here?Adv Parasitol. 2020;108:1-45. doi: 10.1016/bs.apar.2019.12.003. Epub 2020 Mar 4. Adv Parasitol. 2020. PMID: 32291083 Review.
-
Structure-activity relationship and target investigation of 2-aryl quinolines with nematocidal activity.Int J Parasitol Drugs Drug Resist. 2024 Apr;24:100522. doi: 10.1016/j.ijpddr.2024.100522. Epub 2024 Jan 23. Int J Parasitol Drugs Drug Resist. 2024. PMID: 38295619 Free PMC article.
-
Advances in Anthelmintic Target Identification.Int J Mol Sci. 2025 Apr 15;26(8):3738. doi: 10.3390/ijms26083738. Int J Mol Sci. 2025. PMID: 40332360 Free PMC article. Review.
Cited by
-
Mass Spectrometry-Based Proteomics Technologies to Define Endogenous Protein-Protein Interactions and Their Applications to Cancer and Viral Infectious Diseases.Mass Spectrom Rev. 2025 Feb 9:10.1002/mas.21926. doi: 10.1002/mas.21926. Online ahead of print. Mass Spectrom Rev. 2025. PMID: 39924651 Free PMC article. Review.
-
Analysis of Haemonchus embryos at single cell resolution identifies two eukaryotic elongation factors as intervention target candidates.Comput Struct Biotechnol J. 2024 Jan 17;23:1026-1035. doi: 10.1016/j.csbj.2024.01.008. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 38435301 Free PMC article.
-
The Mitogenome of the Haecon-5 Strain of Haemonchus contortus and a Comparative Analysis of Its Nucleotide Variation with Other Laboratory Strains.Int J Mol Sci. 2024 Aug 12;25(16):8765. doi: 10.3390/ijms25168765. Int J Mol Sci. 2024. PMID: 39201452 Free PMC article.
-
Genome-Wide Analysis of Haemonchus contortus Proteases and Protease Inhibitors Using Advanced Informatics Provides Insights into Parasite Biology and Host-Parasite Interactions.Int J Mol Sci. 2023 Aug 1;24(15):12320. doi: 10.3390/ijms241512320. Int J Mol Sci. 2023. PMID: 37569696 Free PMC article.
-
Prediction and Prioritisation of Novel Anthelmintic Candidates from Public Databases Using Deep Learning and Available Bioactivity Data Sets.Int J Mol Sci. 2025 Mar 28;26(7):3134. doi: 10.3390/ijms26073134. Int J Mol Sci. 2025. PMID: 40243899 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

